Mastering the Amplex Red Assay: A Complete Guide to Extracellular H₂O₂ Detection for Biomedical Research
This comprehensive guide details the Amplex Red/horseradish peroxidase (HRP) assay for sensitive, specific detection of extracellular hydrogen peroxide (H₂O₂).
Mastering the Amplex Red Assay: A Complete Guide to Extracellular H₂O₂ Detection for Biomedical Research
Abstract
This comprehensive guide details the Amplex Red/horseradish peroxidase (HRP) assay for sensitive, specific detection of extracellular hydrogen peroxide (H₂O₂). Tailored for researchers and drug development professionals, the article explores the foundational chemistry and biological significance of H₂O₂ signaling, provides step-by-step optimized protocols for diverse in vitro applications, addresses common pitfalls and advanced optimization strategies, and critically evaluates the assay's performance against alternative methods. The synthesis empowers scientists to reliably measure this key reactive oxygen species (ROS) in studies of oxidative stress, redox signaling, inflammation, and drug mechanisms.
H₂O₂ as a Signaling Molecule: Why Detect Extracellular Peroxide with Amplex Red?
Application Notes & Protocols Context: This document supports a thesis investigating the optimization and application of the Amplex Red assay for the specific, sensitive detection of extracellular hydrogen peroxide (H₂O₂) in cellular models, elucidating its dual role in pathophysiology and signaling.
1. Introduction Extracellular H₂O₂ is a key redox-active molecule. At high, sustained concentrations, it contributes to oxidative stress, damaging biomolecules and disrupting tissue homeostasis. At low, transient concentrations, it acts as a deliberate signaling mediator, modulating pathways critical for proliferation, immune response, and differentiation. Precise measurement of its spatiotemporal dynamics is therefore essential. The Amplex Red/Peroxidase assay provides a robust, fluorometric method for real-time quantification of H₂O₂ released into the extracellular milieu.
2. Quantitative Data Summary: H₂O₂ in Physiology & Pathology Table 1: Physiological vs. Pathological Concentrations of Extracellular H₂O₂
| Context / Source | Approximate [H₂O₂] Range | Primary Role | Key Outcome |
|---|---|---|---|
| Basal Cellular Leakage | 10–100 nM | Homeostatic | Low-level background signaling |
| Ligand-Activated Signaling (e.g., EGF, PDGF) | 0.1–1 µM | Redox Signaling | Transient kinase inhibition (e.g., PTPs), gene expression |
| Activated Immune Cell (Neutrophil) Burst | 10–100 µM (local) | Microbial Killing | Oxidative stress on pathogens, potential host tissue damage |
| Chronic Inflammation Site | 1–10 µM (sustained) | Oxidative Stress | DNA/protein/lipid damage, apoptotic signaling, senescence |
| In Vitro Cytotoxicity Studies | 50–500 µM (added) | Induced Oxidative Stress | Modeled cell death (apoptosis/necrosis) |
Table 2: Key Parameters for Amplex Red Assay Optimization
| Parameter | Recommended Condition | Rationale & Impact |
|---|---|---|
| Amplex Red Concentration | 10–50 µM | Balances sensitivity with potential auto-oxidation at high [ ] |
| Horseradish Peroxidase (HRP) | 0.1–0.5 U/mL | Ensures reaction is not HRP-limited; excess can increase background |
| Assay Buffer | HEPES or PBS, pH 7.4 | Maintains physiological pH for HRP activity and cell health during live-cell assays |
| Incubation Temperature | 37°C (live-cell) or RT (cell lysate) | Optimizes enzyme kinetics and reflects biological conditions |
| Detection Mode (Microplate) | Fluorescence: Ex/Em ~560/590 nm | Specific detection of resorufin product; avoid exposure to ambient light. |
| Interference Considerations | Avoid media with phenol red, high antioxidant (ascorbate) levels | Phenol red quenches fluorescence; antioxidants scavenge H₂O₂. |
| Dynamic Range | 10 nM – 50 µM H₂O₂ | Suitable for detecting both signaling and stress-relevant concentrations. |
3. Detailed Protocols
Protocol 3.1: Real-Time Detection of Receptor-Generated Extracellular H₂O₂ using Amplex Red Objective: To quantify ligand-stimulated (e.g., Growth Factor) H₂O₂ production in adherent cell cultures. Materials: Amplex Red reagent, Horseradish Peroxidase (HRP), Hanks' Balanced Salt Solution (HBSS, phenol red-free), target growth factor (e.g., EGF at 100 ng/mL), black-walled clear-bottom 96-well plate, fluorometric microplate reader. Procedure:
- Cell Preparation: Seed cells in plate 24-48h prior to achieve ~80% confluence. On day of assay, wash cells 2x with warm HBSS.
- Reagent Master Mix: Prepare Amplex Red/HRP working solution in HBSS to final concentrations of 50 µM Amplex Red and 0.1 U/mL HRP. Protect from light.
- Baseline Measurement: Add 100 µL/well of working solution. Incubate plate in reader at 37°C for 10 min. Measure fluorescence (Ex/Em 560/590) every minute to establish a stable baseline (3-5 time points).
- Stimulation: At the desired time, briefly pause the reader. Add 10 µL of pre-warmed growth factor solution (prepared at 10x in HBSS) to test wells. Add 10 µL HBSS to control wells. Gently mix by shaking.
- Kinetic Measurement: Immediately resume kinetic fluorescence measurement every 1–2 min for 60–120 min.
- Data Analysis: Subtract average blank (no cells) fluorescence. Plot ΔFluorescence over time. Generate a standard curve using known H₂O₂ concentrations (0-10 µM) in parallel to quantify nmoles of H₂O₂ produced.
Protocol 3.2: Validating H₂O₂ Specificity in Amplex Red Assays Objective: To confirm that the detected signal is specific to H₂O₂. Procedure: Run parallel experiments as in Protocol 3.1 with the following additions:
- Catalase Control: Pre-treat a set of stimulated wells with 1000 U/mL Catalase (a H₂O₂-scavenging enzyme) for 10 min prior to adding Amplex Red/HRP mix. Catalase should abolish >95% of the signal increase.
- HRP-Omission Control: Perform assay without HRP in the working solution. The lack of enzyme should prevent resorufin formation, confirming the peroxidase-dependent nature of the signal.
- Inhibitor Studies: Pre-incubate cells with inhibitors of potential H₂O₂ sources (e.g., 10 µM DPI for NADPH oxidases, 100 µM Allopurinol for xanthine oxidase) for 30 min to probe enzymatic origin.
4. The Scientist's Toolkit: Essential Reagents & Materials Table 3: Key Research Reagent Solutions
| Item | Function & Application Notes |
|---|---|
| Amplex Red (10-Acetyl-3,7-dihydroxyphenoxazine) | Fluorogenic substrate. Reacts with H₂O₂ in a 1:1 stoichiometry via HRP catalysis to yield highly fluorescent resorufin. |
| Recombinant Horseradish Peroxidase (HRP) | Essential enzyme catalyst for the Amplex Red reaction. Must be included in assay buffer. |
| Catalase (from bovine liver) | Negative control reagent. Scavenges H₂O₂; used to confirm signal specificity. |
| Diphenyleneiodonium (DPI) Chloride | Pharmacological inhibitor of flavoprotein enzymes, including many NADPH oxidase (NOX) isoforms. Used to probe source of H₂O₂. |
| Phenol Red-Free Cell Culture Buffer (e.g., HBSS) | Essential assay medium. Phenol red interferes with fluorescence detection at 560/590 nm. |
| Hydrogen Peroxide Standard Solution | Required for generating a standard curve in each experiment to convert fluorescence units to molar concentration. |
| Black-Walled, Clear-Bottom Microplates | Optimizes fluorescence signal by minimizing cross-talk between wells while allowing for microscopic visualization of cells. |
5. Visualization Diagrams
Diagram Title: H₂O₂ Roles and Detection Pathway
Diagram Title: Live-Cell H₂O₂ Detection Protocol
Diagram Title: H₂O₂ Signaling through PTP Oxidation
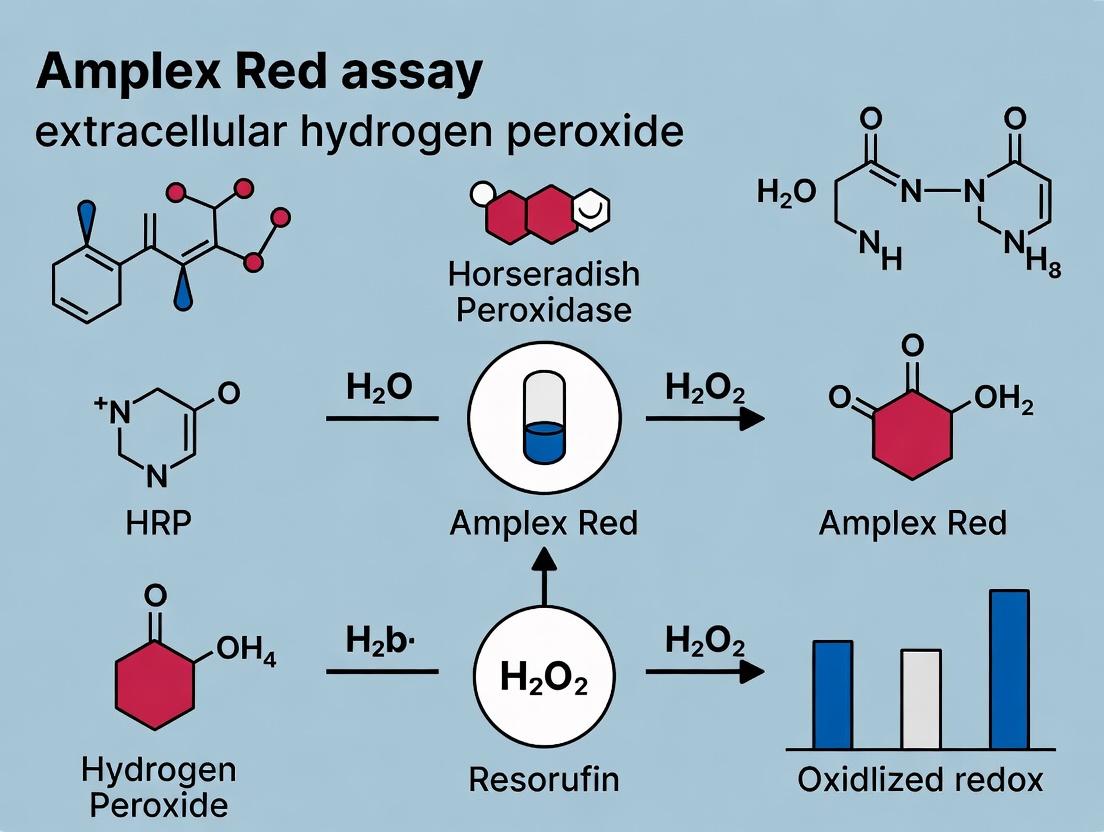
This application note details the core enzymatic chemistry of the Amplex Red assay, a cornerstone method for detecting extracellular hydrogen peroxide (H₂O₂). Within the context of broader research on oxidative stress and redox signaling, understanding the precise catalytic role of Horseradish Peroxidase (HRP) is critical for assay optimization, validation, and data interpretation in drug development and physiological studies.
Core Reaction Mechanism
The Amplex Red assay is a fluorogenic reaction where non-fluorescent Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) is oxidized in the presence of hydrogen peroxide to generate highly fluorescent resorufin. HRP is the essential catalyst that enables this conversion under mild, physiological conditions.
1. Catalytic Cycle of HRP: HRP contains a ferric heme (Fe³⁺) prosthetic group. The catalytic mechanism is a classic peroxidase cycle:
- Step 1: The native Fe³⁺ enzyme reacts with one molecule of H₂O₂, forming the reactive intermediate Compound I (an oxyferryl species, Fe⁴⁺=O, with a porphyrin π-cation radical). This step involves a two-electron oxidation of the enzyme.
- Step 2: Compound I oxidizes one molecule of Amplex Red (the electron donor) to the corresponding radical, itself being reduced to Compound II (oxyferryl species, Fe⁴⁺=O).
- Step 3: Compound II oxidizes a second molecule of Amplex Red, returning the enzyme to its native Fe³⁺ state. The two Amplex Red radicals undergo dismutation to produce the final product, resorufin.
2. Fluorescence Generation: The oxidation and subsequent structural rearrangement of colorless, non-fluorescent Amplex Red yields resorufin, a bright pink dye with strong fluorescence (Ex/Em ~571/585 nm). The fluorescence intensity is directly proportional to the amount of H₂O₂ present in the sample, given that HRP and Amplex Red are in excess.
Table 1: Key Spectral and Kinetic Parameters of the Amplex Red/HRP Reaction
| Parameter | Value | Conditions / Notes |
|---|---|---|
| Amplex Red Absorption Max | ~563 nm | In DMSO or buffer |
| Resorufin Excitation Max | 571 nm | Primary excitation peak |
| Resorufin Emission Max | 585 nm | Primary emission peak |
| Extinction Coefficient (Resorufin) | ~54,000 cm⁻¹M⁻¹ | At 571 nm |
| Assay pH Optimum | 7.4 | Phosphate buffer (range 7.0-8.0) |
| Typical HRP Concentration | 0.1 - 0.2 U/mL | Final reaction concentration |
| Typical Amplex Red Concentration | 50 - 100 µM | Final reaction concentration |
| Detection Limit (H₂O₂) | ~10 - 50 nM | Dependent on instrument sensitivity |
Table 2: Common Interfering Substances & Effects
| Substance | Potential Effect on Amplex Red/HRP Assay | Recommended Action |
|---|---|---|
| Ascorbic Acid | Reduces intermediates, quenches fluorescence; major interference. | Use antioxidant scavengers (e.g., ascorbate oxidase), or purify samples. |
| Thiols (e.g., GSH, DTT) | Can reduce resorufin back to Amplex Red, causing signal decay. | Derivatize or dilute samples to minimize impact. |
| Other Peroxidases | May catalyze the same reaction if present in samples. | Include control reactions without exogenous HRP. |
| Strong Oxidants (e.g., ONOO⁻) | May oxidize Amplex Red non-enzymatically. | Use specific inhibitors or scavengers. |
| HRP Inhibitors (e.g., NaN₃, CN⁻) | Inhibit catalytic activity. | Avoid in assay buffers. |
Detailed Protocol: Measuring Extracellular H₂O₂ from Cultured Cells
Materials & Reagents
The Scientist's Toolkit:
| Item | Function / Specification |
|---|---|
| Amplex Red Reagent | Substrate (10-acetyl-3,7-dihydroxyphenoxazine). Prepare a 10 mM stock in anhydrous DMSO. Aliquot and store at -20°C, protected from light. |
| Horseradish Peroxidase (HRP) | Catalytic enzyme. Use a high-purity, lyophilized powder. Prepare a 100 U/mL stock in reaction buffer. Aliquot and store at -20°C. |
| Reaction Buffer | Typically, pH 7.4. 50 mM sodium phosphate, 100 mM NaCl, or Hanks' Balanced Salt Solution (HBSS) for cell assays. Pre-warm to 37°C. |
| H₂O₂ Standard Solution | For calibration curve. Prepare fresh serial dilutions from a certified 30% stock. Concentration must be verified spectrophotometrically (A₂₄₀, ε=43.6 M⁻¹cm⁻¹). |
| Cell Culture Plates | 96-well or 24-well clear-bottom black plates for fluorescence measurement. |
| Microplate Fluorescence Reader | Equipped with filters or monochromators for ~571 nm excitation and ~585 nm emission. |
| Positive Control | A system known to generate H₂O₂ (e.g., glucose oxidase + glucose, or a phorbol ester-stimulated NADPH oxidase). |
Procedure
- Prepare Working Solution: Combine reaction buffer, Amplex Red stock, and HRP stock to create a working solution with final concentrations of 50-100 µM Amplex Red and 0.1 U/mL HRP. Prepare fresh and protect from light.
- Cell Preparation: Plate cells in a 96-well black plate and grow to desired confluence. On the day of the experiment, wash cells 2x with warm, phenol red-free buffer or HBSS.
- Establish Standard Curve: In empty wells, add the working solution to known concentrations of H₂O₂ (e.g., 0, 0.1, 0.5, 1, 2, 5 µM). Run in triplicate.
- Sample Reaction: Add the Amplex Red/HRP working solution to the washed cells. For background control, add working solution containing a potent HRP inhibitor (e.g., 10 mM sodium azide) to replicate wells.
- Incubation & Measurement: Incubate the plate at 37°C. Measure fluorescence (Ex/Em ~571/585 nm) kinetically (e.g., every 5 minutes for 30-60 minutes) or at a single endpoint (e.g., 30 minutes). Use a top-reading fluorescence plate reader.
- Data Analysis: Subtract the background control (AZIDE) values from sample readings. Calculate H₂O₂ concentrations in sample wells by interpolating from the linear region of the standard curve (fluorescence vs. H₂O₂ concentration).
Critical Protocol Notes
- Light Sensitivity: Protect all reaction mixtures containing Amplex Red from prolonged light exposure.
- Enzyme Stability: Avoid repeated freeze-thaw cycles of HRP and Amplex Red stocks.
- Linearity: The reaction must be performed under conditions where the signal is linear with time and H₂O₂ concentration. Optimize cell number and incubation time.
- Specificity Controls: To confirm the signal is due to H₂O₂, pre-treat samples with catalase (an enzyme that degrades H₂O₂), which should abolish the signal.
Visualizations
Diagram Title: HRP Catalytic Cycle in the Amplex Red Assay
Diagram Title: Amplex Red Assay Protocol Workflow
The detection of extracellular hydrogen peroxide (H₂O₂) is a critical parameter in cell signaling, oxidative stress research, and drug development. The Amplex Red assay, utilizing the horseradish peroxidase (HRP)-catalyzed reaction, has become a cornerstone methodology. Within this broader thesis, its key advantages—superior sensitivity, high specificity, and the capacity for real-time kinetic readouts—define its utility for researchers investigating NADPH oxidase activity, mitochondrial function, and pharmacological modulation of reactive oxygen species (ROS).
Quantitative Performance Data
Table 1: Comparative Performance Metrics of the Amplex Red Assay
| Parameter | Typical Range/Value | Comparative Advantage |
|---|---|---|
| Sensitivity (Detection Limit) | 10–50 nM H₂O₂ | ~10x more sensitive than coupled assays using phenol red or ABTS. |
| Specificity | High for H₂O₂ over other ROS (e.g., superoxide, peroxynitrite). | HRP enzyme provides specificity; minimal interference from superoxide. |
| Linear Dynamic Range | 0.1–10 µM H₂O₂ | Allows quantification of both basal and stimulated cellular production. |
| Assay Time for Kinetics | Continuous, real-time monitoring over minutes to hours. | Enables measurement of rate constants (e.g., Vmax, Km for oxidase activity). |
| Z'-Factor (HTS suitability) | >0.7 in optimized 96-/384-well formats. | Robust for high-throughput drug screening applications. |
Table 2: Interfering Substances & Mitigation Strategies
| Interfering Substance | Effect on Amplex Red Assay | Recommended Mitigation |
|---|---|---|
| Exogenous Antioxidants (e.g., Ascorbate) | Reduces resorufin, causing signal loss. | Include catalase control; use antioxidant scavengers like ascorbate oxidase. |
| HRP Inhibitors (e.g., Azide, Cyanide) | Inhibits core enzymatic reaction. | Avoid in assay buffers; use minimal, consistent wash steps for cells. |
| Cellular Reductants | May non-enzymatically reduce Amplex Red. | Run probe-only controls without HRP. |
| Other Peroxidases | Can produce false-positive signal. | Use specific HRP inhibitors in control wells; use purified HRP in cell-free systems. |
Detailed Protocols
Protocol 1: Standard Calibration and Sensitivity Determination
Objective: Establish a standard curve and determine the lower limit of detection (LLOD) for H₂O₂. Reagents: Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine), Horseradish Peroxidase (HRP, 0.2 U/mL), H₂O₂ standard (from serial dilutions of a 3% stock in reaction buffer), 1X Reaction Buffer (50 mM sodium phosphate, pH 7.4). Procedure:
- Prepare a dilution series of H₂O₂ in buffer (0, 0.05, 0.1, 0.25, 0.5, 1.0, 2.5, 5.0 µM).
- In a black 96-well plate, mix 50 µL of each H₂O₂ standard with 50 µL of working solution (50 µM Amplex Red + 0.2 U/mL HRP in buffer).
- Incubate protected from light at 37°C for 30 minutes.
- Measure fluorescence (Ex/Em = 540/590 nm) using a plate reader.
- LLOD Calculation: LLOD = (3.3 * SD of zero standard) / slope of the linear calibration curve.
Protocol 2: Real-Time Kinetic Measurement from Cultured Cells
Objective: Measure the rate of extracellular H₂O₂ production from adherent cells (e.g., macrophages, endothelial cells) in real-time. Reagents: Complete cell culture medium (phenol-red free), HBSS with Ca²⁺/Mg²⁺, Amplex Red/HRP working solution (as above), Pharmacological agents (e.g., PMA for stimulation, diphenyleneiodonium (DPI) for inhibition). Procedure:
- Plate cells in a clear-bottom black 96-well plate and culture to desired confluence.
- Wash cells twice with warm, phenol-red free HBSS.
- Add 100 µL/well of Amplex Red/HRP working solution in HBSS. Include control wells without cells (background) and without probe (autofluorescence).
- Immediately place plate in a pre-warmed (37°C) fluorescence plate reader.
- Read kinetic fluorescence (Ex/Em = 540/590 nm) every 1-2 minutes for 60-90 minutes to establish baseline.
- At a defined time point (e.g., cycle 10), inject 20 µL of 6X concentrated stimulus (e.g., PMA) or inhibitor using the reader's injector. Continue reading.
- Data Analysis: Subtract background. The rate of H₂O₂ production (nM/min) is calculated from the linear phase of fluorescence increase using the standard curve slope.
Protocol 3: Specificity Validation Using Scavenging Enzymes
Objective: Confirm that the detected signal is specific to H₂O₂. Reagents: Catalase (from bovine liver, 1000 U/mL), Superoxide Dismutase (SOD, 500 U/mL), Cell culture or H₂O₂-generating system (e.g., glucose/glucose oxidase). Procedure:
- Set up experimental wells with cells or a generating system in triplicate.
- Pre-treatment: Add one of the following to separate wells 10 minutes before adding Amplex Red/HRP: a. Catalase (500 U/mL final) – specific H₂O₂ scavenger. b. SOD (250 U/mL final) – converts superoxide to H₂O₂ (may increase signal if superoxide is present). c. Vehicle control.
- Add Amplex Red/HRP working solution and perform kinetic measurement (Protocol 2).
- Interpretation: >90% signal inhibition by catalase confirms specificity for H₂O₂. An increase in signal with SOD indicates precursor superoxide production.
Visualizations
Title: Amplex Red Assay Core Reaction & Kinetic Readout Pathway
Title: Workflow for Real-Time Cell-Based H₂O₂ Detection
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Robust Amplex Red Assays
| Reagent/Material | Function & Critical Notes | Example Vendor/ Cat. # |
|---|---|---|
| Amplex Red Reagent (10-acetyl-3,7-dihydroxyphenoxazine) | Fluorogenic substrate. Specific for HRP in presence of H₂O₂. Light-sensitive; prepare fresh. | Thermo Fisher Scientific, A12222 |
| Horseradish Peroxidase (HRP), Purified | Enzyme catalyst. Critical for reaction specificity and signal amplification. Use consistent lot. | Sigma-Aldrich, P8375 |
| Hydrogen Peroxide, 3% Solution | Primary standard for calibration. Dilute fresh daily from stock for accurate standard curves. | Various |
| Catalase (from bovine liver) | Specificity control. Scavenges H₂O₂; confirms signal origin. High-purity grade recommended. | Sigma-Aldrich, C9322 |
| Phenol Red-Free Cell Culture Medium | Eliminates background fluorescence and pH interference during kinetic reads. | Gibco, 21063029 |
| Black/Clear-Bottom 96- or 384-Well Plates | Optimal for fluorescence measurement while allowing microscopic visualization of cells. | Corning, 3603 |
| Diphenyleneiodonium (DPI) Chloride | Flavoprotein inhibitor (e.g., inhibits NADPH oxidases). Key negative control for cellular assays. | Tocris, 2638 |
| Phorbol 12-Myristate 13-Acetate (PMA) | Protein kinase C activator; potent stimulator of NADPH oxidase in immune cells. | Sigma-Aldrich, P8139 |
| Microplate Reader with Kinetic Capability | Must have temperature control (37°C), injectors, and appropriate filters (∼540/590 nm). | Instruments from BMG Labtech, BioTek, etc. |
Application Note 1: Cell Signaling Pathway Modulation
In the context of the broader thesis on the Amplex Red assay, this methodology is pivotal for quantifying extracellular H₂O₂ released during specific receptor-mediated signaling events. The assay provides a real-time, sensitive readout of reactive oxygen species (ROS) production, a key secondary messenger in pathways like EGFR and NOD2.
Protocol: Measuring Receptor-Mediated H₂O₂ Burst in Adherent Cells (e.g., EGFR Activation)
- Day 1: Cell Seeding. Seed adherent cells (e.g., A431, HeLa) at 80% confluence in a clear-bottom 96-well plate. Use phenol red-free culture medium.
- Day 2: Assay Setup.
- Prepare 1X Reaction Buffer: 50 mM sodium phosphate buffer, pH 7.4, containing 138 mM NaCl, 2.7 mM KCl.
- Prepare Amplex Red Working Solution: Dilute Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine) in 1X Reaction Buffer to a final concentration of 50 µM. Add Horseradish Peroxidase (HRP) to a final concentration of 0.1 U/mL.
- Wash cells 2x with 100 µL of warm, sterile 1X Reaction Buffer.
- Add 90 µL of Amplex Red/HRP Working Solution to each well.
- Add 10 µL of agonist (e.g., EGF at 100 ng/mL final concentration) or inhibitor (e.g., AG1478) prepared in Reaction Buffer to appropriate wells. Include vehicle controls.
- Immediately place plate in a pre-warmed (37°C) fluorescence microplate reader.
- Measurement: Measure fluorescence (Ex/Em = 530-560 nm / 580-610 nm, with a cut-off ~590 nm) every 5 minutes for 60-120 minutes.
- Quantification: Generate a standard curve using known concentrations of H₂O₂ (0 to 20 µM) in parallel. Calculate extracellular H₂O₂ concentration from fluorescence values.
Table 1: Quantified H₂O₂ Production from EGFR Signaling
| Cell Line | Stimulus | Inhibitor | Max H₂O₂ Accumulation (µM, mean ± SD) | Time to Peak (min) |
|---|---|---|---|---|
| A431 | EGF (100 ng/mL) | None | 8.2 ± 0.9 | 45 |
| A431 | EGF (100 ng/mL) | AG1478 (10 µM) | 1.1 ± 0.3 | - |
| HeLa | EGF (100 ng/mL) | None | 3.5 ± 0.6 | 60 |
| HeLa | Serum-Free Media | None | 0.5 ± 0.2 | - |
Application Note 2: High-Throughput Drug Screening
The Amplex Red assay is optimized for high-throughput screening (HTS) of compound libraries for modulators of cellular ROS metabolism, identifying potential antioxidants or pro-oxidant therapeutics.
Protocol: 384-Well HTS for NOX4 Inhibitors
- Automated Cell Seeding: Seed NOX4-expressing cells (e.g., HEK293-NOX4) in 384-well black-walled, clear-bottom plates at 5,000 cells/well in 40 µL growth medium. Incubate overnight.
- Compound Addition: Using a pin tool or liquid handler, transfer 100 nL of test compounds from a 10 mM DMSO stock library into wells (final compound concentration ~10 µM). Include controls (DMSO vehicle, positive control inhibitor like GKT137831).
- Amplex Red Addition: After 30 min pre-incubation, add 20 µL of 3X concentrated Amplex Red/HRP Working Solution (final: 50 µM Amplex Red, 0.1 U/mL HRP) using a multidispenser.
- Kinetic Read: Immediately measure fluorescence intensity every 2 minutes for 60 minutes at 37°C.
- Data Analysis: Calculate the initial rate of fluorescence increase (RFU/min) for each well between 5-20 minutes. Normalize to vehicle control (100% activity) and positive inhibitor (0% activity). Compounds showing >70% inhibition are considered primary hits.
Table 2: HTS Results from a NOX4 Inhibitor Screen
| Plate | Total Wells | Vehicle Control (RFU/min) | Positive Inhibitor (RFU/min) | Primary Hits (>70% Inhib) | Z'-Factor |
|---|---|---|---|---|---|
| 1 | 384 | 125 ± 8 | 22 ± 4 | 12 | 0.78 |
| 2 | 384 | 118 ± 10 | 20 ± 3 | 9 | 0.81 |
| Total | 768 | 121.5 ± 9.3 | 21 ± 3.5 | 21 | 0.79 |
Application Note 3: Inflammatory Response Modeling
This application leverages the Amplex Red assay to model and quantify the oxidative burst from immune cells (e.g., macrophages) in response to inflammatory stimuli, such as LPS or cytokines.
Protocol: Measuring Macrophage Oxidative Burst
- Cell Preparation: Differentiate THP-1 monocytes into macrophages using 100 nM PMA for 48 hours, followed by 24-hour rest in RPMI with 10% FBS. Use primary murine BMDMs as an alternative.
- Assay Execution:
- Wash adherent macrophages 2x with Krebs-Ringer Phosphate (KRP) buffer.
- Add 90 µL of Amplex Red/HRP Working Solution in KRP buffer.
- Stimulate cells by adding 10 µL of LPS (1 µg/mL final) ± IFN-γ (20 ng/mL final) or vehicle.
- Measure fluorescence kinetically (Ex/Em ~571/585 nm) for 2-4 hours at 37°C.
- Advanced Model: For a co-culture inflammatory model, seed macrophages in transwell inserts, stimulate, and measure Amplex Red signal in the lower chamber containing endothelial cells to quantify paracrine H₂O₂ signaling.
Table 3: H₂O₂ Production in Inflammatory Models
| Cell Type | Stimulus | Amplex Red Signal (Fold Increase vs. Untreated) | Significance (p-value) |
|---|---|---|---|
| THP-1 Macrophage | None (Basal) | 1.0 ± 0.2 | - |
| THP-1 Macrophage | LPS (1 µg/mL) | 3.8 ± 0.5 | <0.001 |
| THP-1 Macrophage | LPS + IFN-γ | 6.4 ± 0.9 | <0.001 |
| Murine BMDM | LPS (1 µg/mL) | 4.2 ± 0.7 | <0.001 |
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function in Amplex Red Assay |
|---|---|
| Amplex Red Reagent | The core substrate. In the presence of HRP and H₂O₂, it is oxidized to fluorescent resorufin. |
| Horseradish Peroxidase (HRP) | Enzyme that catalyzes the oxidation of Amplex Red by H₂O₂. Essential for reaction. |
| Phenol Red-Free Medium | Eliminates background fluorescence and potential interference from pH-sensitive dyes. |
| Sodium Pyruvate | Often added to culture medium to scavenge endogenous H₂O₂, reducing basal signal. |
| Catalase | Negative control enzyme. Specifically degrades H₂O₂, confirming signal specificity. |
| DPI (Diphenyleneiodonium) | Broad NADPH oxidase (NOX) inhibitor. Used as a pharmacological control for cellular H₂O₂ production. |
| H₂O₂ Standard Solution | Used to generate a standard curve for quantitative conversion of fluorescence to µM H₂O₂. |
| HRP-Conjugated Antibodies | Potential source of contamination in cell-based assays; use HRP-free alternatives for immunostaining prior to assay. |
Title: H₂O₂ Detection in EGFR Signaling Pathway
Title: High-Throughput Drug Screening Protocol Flow
Title: Macrophage Inflammatory Signaling to H₂O₂
Step-by-Step Protocol: Optimizing Amplex Red for Your Experimental System
Reagent Preparation and Critical Storage Conditions for Stable Results
Within the thesis "Quantitative Dynamics of Extracellular Hydrogen Peroxide in Drug-Treated Cancer Cell Lines Using the Amplex Red Assay," the reliability of data is fundamentally dependent on precise reagent preparation and stringent storage. The Amplex Red/Peroxidase system is highly sensitive to environmental factors; degradation of key components leads to increased background fluorescence and diminished sensitivity, compromising research conclusions on H₂O₂ flux. This document details standardized protocols and critical storage parameters to ensure assay stability and reproducibility.
Key Reagents and Their Functions
Table 1: Research Reagent Solutions for Amplex Red Assay
| Reagent | Function | Critical Storage Parameter |
|---|---|---|
| Amplex Red (10-Acetyl-3,7-dihydroxyphenoxazine) | Probe molecule. Enzymatically oxidized by HRP in the presence of H₂O₂ to generate highly fluorescent resorufin. | -20°C, desiccated, dark. Aliquot to avoid freeze-thaw cycles. Stable for ~6 months. |
| Horseradish Peroxidase (HRP) | Enzyme catalyst. Drives the oxidation of Amplex Red by H₂O₂. | -20°C. Avoid repeated freezing/thawing; use working aliquots. |
| Reaction Buffer (e.g., Krebs-Ringer Phosphate) | Provides physiologically relevant ionic milieu for extracellular detection. | 4°C. Check pH before each use (recommended pH 7.4). |
| H₂O₂ Standard Stock | Primary calibrant for standard curve generation. | -20°C in small, single-use aliquots. Concentration must be verified spectrophotometrically (ε₂₄₀ = 43.6 M⁻¹cm⁻¹). |
| DMSO (Cell Culture Grade) | Solvent for preparing Amplex Red stock solution. | Room temperature, anhydrous. |
| Catalase | Negative control enzyme; specifically scavenges H₂O₂. | -20°C. |
Detailed Preparation Protocols
Primary Stock Solutions
Amplex Red Stock Solution (20 mM in DMSO):
- Warm Amplex Red vial to room temperature before opening.
- Add the recommended volume of anhydrous DMSO to achieve a 20 mM concentration.
- Vortex gently until fully dissolved.
- Immediately aliquot (e.g., 50 µL) into single-use, light-protected microcentrifuge tubes.
- Store at -20°C in a desiccator, protected from light. Discard any aliquot after use.
Horseradish Peroxidase (HRP) Stock Solution (100 U/mL in buffer):
- Centrifuge the lyophilized powder vial briefly before opening.
- Reconstitute in ice-cold reaction buffer (not water) to 100 U/mL.
- Gently mix by inversion. Do not vortex vigorously.
- Prepare small aliquots (e.g., 20 µL) and store at -20°C.
H₂O₂ Standard Stock (10 mM in buffer):
- Dilute a fresh, concentrated H₂O₂ solution (e.g., 30%) in ice-cold reaction buffer.
- Determine exact concentration by absorbance at 240 nm (A₂₄₀ = εcl).
- Prepare a 10 mM stock based on the calculated concentration.
- Aliquot and store at -20°C. Use within one week for critical work.
Working Solution Preparation
- Amplex Red/HRP Working Solution:
- Thaw reagents on ice.
- Prepare fresh for each experiment in a light-protected tube.
- Final typical concentration: 100 µM Amplex Red, 0.2 U/mL HRP in reaction buffer.
- Example for 10 mL: Add 50 µL of 20 mM Amplex Red stock + 20 µL of 100 U/mL HRP stock to 9.93 mL of pre-warmed (37°C) reaction buffer.
- Mix by gentle inversion and keep in the dark at 37°C until use. Discard after 2 hours.
Quantitative Stability Data
Table 2: Impact of Storage Conditions on Reagent Performance
| Reagent | Condition Tested | Key Metric | Result | Recommended Max Storage |
|---|---|---|---|---|
| Amplex Red Working Solution (100 µM) | 37°C, exposed to ambient light | Background Fluorescence (RFU) | 450% increase after 4 hours | Use immediately; discard after 2 hrs |
| Amplex Red Stock (20 mM in DMSO) | -20°C, desiccated vs. -20°C, non-desiccated | Assay Signal-to-Noise Ratio | 25% loss in S/N after 3 months (non-desiccated) | 6 months (desiccated, dark, -20°C) |
| HRP Stock (100 U/mL) | 4°C vs. -20°C aliquots | Enzymatic Activity (% remaining) | ~60% activity after 1 week at 4°C | 1 month at -20°C (aliquoted) |
| H₂O₂ Std (10 mM in buffer) | 4°C vs. -20°C aliquots | Concentration Accuracy (% of initial) | ~40% degradation after 1 week at 4°C | 1 week at -20°C |
Experimental Protocol: H₂O₂ Detection from Adherent Cells
Title: Amplex Red Assay for Extracellular H₂O₂ Detection. Materials: Prepared working solution, cell culture plate, H₂O₂ standards, microplate reader capable of fluorescence detection (Ex/Em ~571/585 nm). Procedure:
- Plate Cells: Seed cells in a clear-bottom 96-well plate. Include cell-free wells for background.
- Prepare Standard Curve: In cell-free wells, create a dilution series of H₂O₂ standard (e.g., 0 to 10 µM) in reaction buffer.
- Aspirate & Wash: On experiment day, aspirate cell culture medium. Gently wash cells 2x with warm PBS or reaction buffer.
- Add Working Solution: Add 100 µL of freshly prepared Amplex Red/HRP working solution to all wells (samples, standards, blanks).
- Incubate & Measure: Immediately place plate in a pre-warmed (37°C) microplate reader. Measure fluorescence kinetically every 5 minutes for 60-90 minutes.
- Data Analysis: Subtract blank (no H₂O₂) values. Generate standard curve from H₂O₂ standard wells. Calculate unknown H₂O₂ concentrations from the linear regression of the standard curve.
Visualization: Signaling Pathways and Workflow
Title: Amplex Red Detection of Cell-Derived Hydrogen Peroxide
Title: Amplex Red Assay Workflow for Stable Results
Application Notes
This document details an optimized, end-to-end protocol for plate-based assays, with specific application within a thesis research project focused on the Amplex Red assay for the detection of extracellular hydrogen peroxide (H₂O₂). This workflow ensures reproducibility, minimizes variability, and is designed for high-throughput screening of compounds that modulate H₂O₂ production in adherent cell cultures. The Amplex Red assay utilizes horseradish peroxidase (HRP) to catalyze the reaction between H₂O₂ and the non-fluorescent Amplex Red reagent, producing the highly fluorescent resorufin, enabling sensitive quantification of extracellular H₂O₂ flux.
1. Experimental Protocol: Optimized End-to-End Workflow
A. Cell Seeding and Culture (Day 1) Objective: To achieve uniform, adherent monolayers with consistent cell density across all wells of a multi-well plate.
- Trypsinization & Counting: Harvest cells using standard trypsin-EDTA procedure. Neutralize trypsin with complete growth medium. Perform an accurate cell count using an automated cell counter or hemocytometer.
- Cell Suspension Preparation: Dilute the cell stock to the optimal seeding density in pre-warmed complete growth medium. Critical: Determine density empirically to ensure 70-90% confluency at assay time (e.g., 10,000 - 50,000 cells/well for a 96-well plate).
- Seeding: Using a multichannel pipette or automated dispenser, aliquot the cell suspension into the inner 60 wells of a clear-bottom, black-walled 96-well plate. The outer perimeter wells receive 100-200 µL of PBS or medium only to minimize evaporation edge effects.
- Incubation: Gently move the plate in a figure-eight motion on the benchtop to ensure even distribution. Place in a humidified 37°C, 5% CO₂ incubator for 24 hours (or until desired confluency is reached).
B. Compound Treatment & Stimulation (Day 2) Objective: To expose cells to experimental modulators (e.g., drug candidates, pathway agonists/antagonists) and/or stimulators of H₂O₂ production.
- Preparation of Compound Plates: In a separate U-bottom or V-bottom plate, perform serial dilutions of test compounds in assay buffer (e.g., Krebs-Ringer Phosphate buffer) or serum-free medium.
- Cell Washing: Gently aspirate the growth medium from the assay plate using a multichannel aspirator. Wash cells once with 100 µL pre-warmed, serum-free assay buffer.
- Compound Addition: Using a multichannel pipette, transfer 90 µL of the compound solutions from the dilution plate to the corresponding wells of the assay plate. Incubate for the desired pre-treatment time (e.g., 30-60 minutes).
- Stimulator Addition: If applicable, prepare a stimulator (e.g., phorbol 12-myristate 13-acetate (PMA) for NADPH oxidase activation) in assay buffer. Add 10 µL directly to wells for a 10X concentrated solution, yielding final desired stimulation volume. Gently mix by orbital shaking.
C. Amplex Red Reaction & Fluorescence Measurement Objective: To initiate the enzymatic detection of extracellular H₂O₂ and capture kinetic fluorescence data.
- Amplex Red/HRP Working Solution: Prepare the reaction mix immediately before use, protected from light. Final concentrations in wells typically are: 50 µM Amplex Red reagent and 0.1 U/mL HRP in assay buffer.
- Reaction Initiation: At time=0, add 100 µL of the Amplex Red/HRP working solution to each well using a multichannel pipette or reagent dispenser. Final total assay volume is 200 µL.
- Immediate Plate Reading: Quickly place the plate in a pre-warmed (37°C) microplate reader.
- Fluorescence Measurement:
- Mode: Kinetic fluorescence measurement.
- Excitation/Emission: 530-540 nm / 580-590 nm (e.g., Ex: 535 nm, Em: 590 nm).
- Measurement Interval: Every 2-5 minutes for 60-120 minutes.
- Gain: Set automatically or manually based on control well signal to avoid saturation.
- Orbital Shaking: Shake for 3-5 seconds before each read to ensure mixing and gas equilibrium.
D. Data Analysis
- Baseline Correction: Subtract the average fluorescence value of time=0 for each well from all subsequent time points for that well.
- Slope Calculation: For each well, calculate the linear rate of fluorescence increase (ΔF/min) over the initial linear phase (typically first 30-60 minutes).
- Normalization: Normalize ΔF/min rates to relevant controls (e.g., untreated cells, vehicle control, or maximum stimulator response).
- H₂O₂ Quantification: Generate a standard curve using known concentrations of H₂O₂ (0 to 10 µM) processed identically alongside experimental wells. Convert ΔF/min to pmol/min H₂O₂ production rate.
2. Quantitative Data Summary
Table 1: Typical H₂O₂ Standard Curve Data for Amplex Red Assay
| H₂O₂ Standard (µM) | Mean Fluorescence (RFU) at t=30 min | Slope (ΔRFU/min) |
|---|---|---|
| 0 | 150 ± 20 | 0.5 ± 0.2 |
| 1 | 1250 ± 150 | 38 ± 4 |
| 2 | 2350 ± 200 | 75 ± 6 |
| 5 | 5750 ± 350 | 188 ± 10 |
| 10 | 11500 ± 500 | 375 ± 15 |
Table 2: Optimized Parameters for Key Workflow Steps
| Step | Parameter | Optimized Value / Recommendation | Purpose |
|---|---|---|---|
| Cell Seeding | Plate Type | Black-walled, clear-bottom 96-well | Minimizes crosstalk, allows microscopy check |
| Seeding Uniformity | CV < 10% (cell count) | Reduces well-to-well variability | |
| Amplex Red Reaction | Final [HRP] | 0.1 U/mL | Ensures reaction is not HRP-limited |
| Final [Amplex Red] | 50 µM | Balances sensitivity and cost | |
| Plate Reading | Temperature Control | 37°C maintained | Preserves physiological enzyme kinetics |
| Read Interval | 3 minutes | Captures kinetics without photobleaching |
3. Diagrams
Optimized Plate-Based Assay Workflow
Amplex Red Detection of NADPH Oxidase-Derived H₂O₂
4. The Scientist's Toolkit: Essential Research Reagent Solutions
| Item / Reagent | Function in the Workflow |
|---|---|
| Amplex Red Reagent (10-Acetyl-3,7-dihydroxyphenoxazine) | The core probe. Enzymatically oxidized by H₂O₂ in the presence of HRP to produce fluorescent resorufin. |
| Horseradish Peroxidase (HRP) | Enzyme catalyst for the Amplex Red reaction. Must be present in excess to ensure reaction rate is limited by H₂O₂ concentration. |
| Cell Culture-Tested 96-Well Plate (Black, clear bottom) | Minimizes optical crosstalk between wells (black walls) while allowing visual inspection of cell monolayers (clear bottom). |
| Krebs-Ringer Phosphate (KRP) Buffer | Common physiological assay buffer that provides ions and pH stability without interference from serum components (e.g., catalase). |
| Phorbol 12-Myristate 13-Acetate (PMA) | A potent pharmacological stimulator of classical NADPH oxidase isoforms, used as a positive control to induce robust H₂O₂ production. |
| Catalase (from bovine liver) | Critical negative control enzyme. Specifically scavenges H₂O₂; addition to assay wells should abolish the fluorescence signal, confirming its specificity. |
| Dimethyl Sulfoxide (DMSO), cell culture grade | Universal solvent for many lipophilic compounds and stimulators (e.g., PMA). Vehicle controls must match the final DMSO concentration in test wells (typically ≤0.1%). |
| Hydrogen Peroxide Standard Solution | Used to generate a standard curve for converting fluorescence slope (ΔRFU/min) into absolute H₂O₂ production rates (pmol/min). |
Application Note: This document details specific methodological adaptations for the Amplex Red hydrogen peroxide (H₂O₂) detection assay, framed within a thesis investigating extracellular H₂O₂ flux in diverse biological systems. Accurate quantification requires protocol optimization for cell type (adherent vs. suspension), sample type (conditioned media), and enzymatic sources.
I. Protocol for Adherent Mammalian Cells
Objective: To measure H₂O₂ released from adherent cell monolayers (e.g., HEK293, HeLa, primary fibroblasts) in real-time.
Key Reagents & Considerations:
- Cell Health: Assay reagents are non-toxic, allowing for continuous kinetic measurement.
- Serum Interference: Phenol red and antioxidants in serum can quench signal. Use reduced serum (≤0.5%) or serum-free, buffered media (e.g., HBSS) during the assay.
- Probe Loading: Amplex Red and Horseradish Peroxidase (HRP) are added directly to the culture well.
Detailed Protocol:
- Plate cells in a clear-bottom, 96-well plate and grow to desired confluency (typically 70-90%).
- Prior to assay, gently wash cells twice with warm, serum-free buffer (e.g., Krebs-Ringer Phosphate buffer, pH 7.4).
- Prepare a working solution of Amplex Red (100 µM) and HRP (0.2 U/mL) in pre-warmed, serum-free, phenol red-free assay buffer.
- Remove wash buffer and immediately add 100 µL/well of the Amplex Red/HRP working solution.
- For background control wells, add working solution plus a high concentration of catalase (500 U/mL).
- Incubate plate at 37°C for 10-30 min to establish a stable baseline fluorescence (Ex/Em ~560/590 nm).
- Apply experimental treatments (e.g., growth factors, drugs, stressors) directly to wells. Mix gently by orbital shaking.
- Record fluorescence kinetically for 60-120 minutes.
- Calculate net H₂O₂ production by subtracting the average background control (Catalase) value and interpolating from an H₂O₂ standard curve run in parallel under identical buffer conditions.
II. Protocol for Suspension Cells
Objective: To measure H₂O₂ released from cells in suspension (e.g., leukocytes, lymphocytes, yeast).
Key Reagents & Considerations:
- Cell Sedimentation: Continuous mixing is required to prevent cell settling, which causes signal fluctuation.
- Cell Number: Optimal signal is highly dependent on cell density; titration is required.
- Centrifugation: May be needed to wash cells free of catalase/antioxidants present in growth media.
Detailed Protocol:
- Harvest suspension cells and wash twice by gentle centrifugation (300 x g, 5 min) in assay buffer.
- Resuspend cells to a density of 0.5-2.0 x 10⁶ cells/mL in assay buffer. Keep on ice.
- Aliquot 90 µL of cell suspension per well into a 96-well plate.
- Prepare a 10X concentrated Amplex Red/HRP working solution (1 mM Amplex Red, 2 U/mL HRP in assay buffer).
- Add 10 µL of the 10X probe solution to each well to achieve final concentrations of 100 µM and 0.2 U/mL, respectively. Mix gently.
- Include background control wells with cells + probe + catalase (500 U/mL final).
- Place plate in pre-warmed microplate reader. Incubate with orbital shaking (1-2 mm circular, medium frequency) between reads.
- After a 10-min baseline read, pause the reader, add treatments (in a minimal volume, e.g., 1-5 µL), resume shaking, and continue kinetic measurement.
- Analyze data as for adherent cells, normalizing final H₂O₂ values to cell number if required.
III. Protocol for Conditioned Media Analysis
Objective: To quantify cumulative H₂O₂ accumulated in cell culture media over a defined period.
Key Reagents & Considerations:
- No Live Cells: The assay measures stable H₂O₂ that has accumulated.
- Catalase/Peroxidase in Media: Fetal bovine serum contains catalase. Must use serum-free conditioned media or treat samples with azide to inhibit endogenous peroxidases.
- Sensitivity: Requires longer incubation as H₂O₂ concentration may be low.
Detailed Protocol:
- Culture cells under experimental conditions. At collection time, carefully aspirate media and centrifuge (1000 x g, 5 min) to remove any detached cells/debris.
- Transfer clarified conditioned media to a new tube. Keep on ice. Assay immediately or snap-freeze for later analysis (single freeze-thaw cycle).
- Prepare a master mix of Amplex Red (100 µM final) and HRP (0.2 U/mL final) in a clean buffer.
- In a 96-well plate, combine 50 µL of conditioned media with 50 µL of the Amplex Red/HRP master mix.
- Include controls: a) Media-only + master mix (media background), b) Conditioned media + master mix + catalase (500 U/mL) (specificity control), c) H₂O₂ standards in fresh, serum-free media.
- Incubate the reaction at 37°C protected from light for 30-60 minutes. Do not use kinetic mode.
- Measure endpoint fluorescence (Ex/Em ~560/590 nm).
- Subtract the average media-only background value from all readings. Calculate H₂O₂ concentration using the standard curve, factoring in any sample dilution.
IV. Protocol for Enzyme Reactions (e.g., NADPH Oxidase, Xanthine Oxidase)
Objective: To measure H₂O₂ production by purified or semi-purified enzyme systems.
Key Reagents & Considerations:
- Direct Measurement: No cellular complexity; defines maximal assay sensitivity.
- Enzyme Cofactors: Must provide necessary substrates (e.g., NADPH, xanthine).
- Inhibitor Controls: Validate signal specificity with enzyme-specific inhibitors (e.g., diphenyleneiodonium for NOX, allopurinol for XO).
Detailed Protocol (NADPH Oxidase Example):
- Prepare a reaction buffer (e.g., 50 mM phosphate buffer, pH 7.0, with 100 µM EGTA).
- In a 96-well plate, combine:
- Buffer
- Enzyme source (e.g., NOX isoform, cell membrane fraction)
- Amplex Red (50 µM final)
- HRP (0.1 U/mL final)
- Substrate (e.g., 100 µM NADPH)
- Set up control wells lacking substrate (background) and lacking enzyme (reagent background). Include an H₂O₂ standard curve in buffer.
- Initiate the reaction by adding the substrate. Mix immediately.
- Record fluorescence kinetically at 30-second intervals for 30-60 minutes at 37°C.
- Calculate initial reaction rates (RFU/min) from the linear phase and convert to pmol/min/mL using the standard curve.
Table 1: Key Assay Parameters for Different Sample Types
| Sample Type | Amplex Red [Final] | HRP [Final] | Incubation Time | Key Interference | Primary Control |
|---|---|---|---|---|---|
| Adherent Cells | 50-100 µM | 0.1-0.2 U/mL | Kinetic (60-120 min) | Serum, Phenol Red | Catalase (in-well) |
| Suspension Cells | 50-100 µM | 0.1-0.2 U/mL | Kinetic (60-120 min) | Cell Settling | Catalase (in-well) |
| Conditioned Media | 50-100 µM | 0.2 U/mL | Endpoint (30-60 min) | Serum Catalase | Catalase + Media Blank |
| Enzyme Reaction | 10-50 µM | 0.1 U/mL | Kinetic (30-60 min) | Substrate Auto-oxidation | Minus Substrate/Enzyme |
Table 2: Expected H₂O₂ Detection Ranges & Limits
| System | Typical Baseline | Stimulated Range | Lower Limit of Detection* | Assay Linear Range* |
|---|---|---|---|---|
| Cell Culture (per 10⁵ cells) | 10-50 pmol | 50-1000 pmol/hr | ~50 nM | 0.1 - 50 µM |
| Conditioned Media | ND - 100 nM | 0.1 - 5 µM | ~50 nM | 0.1 - 50 µM |
| Purified Enzyme | N/A | Varies by activity | ~10 nM | 0.01 - 50 µM |
*Dependent on instrument sensitivity and background. Values are typical for plate readers.
VI. The Scientist's Toolkit: Essential Reagents & Materials
| Item | Function & Rationale |
|---|---|
| Amplex Red (10-Acetyl-3,7-dihydroxyphenoxazine) | Nearly non-fluorescent probe that reacts with H₂O₂ in a 1:1 stoichiometry via HRP to yield fluorescent resorufin. |
| Horseradish Peroxidase (HRP) | Enzyme catalyst for the oxidation of Amplex Red by H₂O₂. Essential for reaction. |
| Catalase (from bovine liver) | Positive control. Rapidly degrades H₂O₂ to H₂O and O₂, confirming signal specificity. |
| Hydrogen Peroxide (30% stock) | Used to generate a standard curve for absolute quantification. Must be freshly diluted. |
| Phenol Red-Free, Serum-Free Buffer | Assay buffer (e.g., HBSS, KRP) to remove serum antioxidants and fluorescent interference. |
| Dimethyl Sulfoxide (DMSO, anhydrous) | High-quality solvent for preparing Amplex Red stock solutions (typically 10-20 mM). |
| Clear-Bottom 96-Well Microplate | Optically clear for fluorescence bottom-reading. Tissue-culture treated for adherent cells. |
| Fluorescence Microplate Reader | Equipped with filters/optics for ~560 nm excitation and ~590 nm emission. Temperature control and kinetic software required. |
| Sodium Azide | Inhibitor of heme peroxidases (including HRP). Used to confirm signal is peroxidase-dependent in complex samples. |
| Diphenyleneiodonium (DPI) | Flavoprotein inhibitor. Used as a negative control in cellular systems to implicate NADPH oxidases as the H₂O₂ source. |
VII. Visualization of Protocols and Pathways
Amplex Red Core Reaction Pathway
Protocol Selection & Validation Workflow
This guide, framed within a broader thesis on the Amplex Red assay for extracellular hydrogen peroxide (H₂O₂) detection, details instrumentation best practices for obtaining reliable, high-quality fluorescence data. Accurate quantification of H₂O₂ is critical in oxidative stress research, signaling studies, and drug development. The performance of fluorometers and microplate readers directly impacts assay sensitivity, reproducibility, and dynamic range.
Quantitative Performance Metrics for Instrument Selection
Selecting appropriate instrumentation requires evaluating key performance parameters. The following table summarizes critical specifications for optimal Amplex Red assay execution.
Table 1: Key Instrument Specifications for Sensitive Fluorescence Assays (e.g., Amplex Red)
| Parameter | Recommended Specification | Impact on Amplex Red Assay |
|---|---|---|
| Detection Mode | Top-read fluorescence (for cell-based assays) or bottom-read | Minimizes interference from cells or particulates in suspension. |
| Excitation/Emission | Filter-based or monochromator (Ex: ~530-570 nm, Em: ~580-610 nm) | Precise targeting of resorufin fluorescence (λmax Ex/Em ~571/585 nm). |
| Sensitivity (for Resorufin) | ≤ 1 pM (in low fluorescence black plates) | Enables detection of low, physiologically relevant H₂O₂ fluxes. |
| Dynamic Range | ≥ 4 orders of magnitude | Accommodates wide range of H₂O₂ concentrations from baseline to stimulated release. |
| Well-to-Well Crosstalk | < 0.1% | Prevents signal bleed between adjacent wells, crucial for 96- or 384-well formats. |
| Temperature Control | Ambient +5°C to 45°C, ±0.5°C accuracy | Essential for maintaining consistent enzyme (HRP) kinetics and cellular activity. |
| Atmospheric Control | CO₂/O₂ control (for live-cell assays) | Maintains physiological pH and health in long-term kinetic measurements. |
Detailed Experimental Protocol: Amplex Red Assay for Extracellular H₂O₂
Adapted from current methodologies for drug screening and oxidative stress research.
A. Reagent Preparation
- Amplex Red Stock Solution (10 mM): Dissolve 5 mg of Amplex Red reagent (N-Acetyl-3,7-dihydroxyphenoxazine) in 1.56 mL of anhydrous DMSO. Aliquot and store at ≤ -20°C, protected from light and moisture. Thaw aliquots on ice.
- Working Solution (100 µM Amplex Red / 0.2 U/mL HRP): Dilute the stock solution 1:100 in 1X reaction buffer (e.g., Krebs-Ringer phosphate buffer, pH 7.4) to 100 µM. Add horseradish peroxidase (HRP) to a final concentration of 0.2 U/mL. Prepare fresh for each experiment and keep on ice, protected from light.
B. Cell-Based Assay Protocol (96-well plate)
- Cell Seeding: Seed adherent cells in a clear-bottomed, black-walled 96-well microplate at desired density. Include cell-free wells for background correction. Culture until confluent or desired state.
- Pre-read & Calibration: Optional: Perform a fluorescence baseline read (Ex/Em ~571/585 nm) to account for background. A resorufin standard curve (0 nM to 10 µM) can be run in parallel for quantification.
- Assay Execution: Carefully aspirate growth medium. Gently wash cells twice with warm, assay-compatible buffer (e.g., HBSS).
- Reaction Initiation: Add 100 µL of the freshly prepared Amplex Red/HRP working solution to each well. For inhibitor/drug studies, pre-incubate cells with compounds for specified time before adding working solution.
- Kinetic Measurement: Immediately place plate in pre-warmed (37°C) reader. Perform kinetic fluorescence measurements every 1-5 minutes for 30-120 minutes, using optimal gain settings determined from control wells.
- Termination & Data Analysis: The reaction is continuous. Data is analyzed as the rate of fluorescence increase (RFU/min) over the linear period, subtracting the background rate from no-cell or negative control wells.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents for Amplex Red-based H₂O₂ Detection Research
| Reagent/Material | Function & Critical Notes |
|---|---|
| Amplex Red Reagent | Nearly non-fluorescent substrate oxidized by HRP in the presence of H₂O₂ to highly fluorescent resorufin. Must be stored anhydrous and protected from light. |
| Horseradish Peroxidase (HRP) | Enzyme that catalyzes the oxidation of Amplex Red. Use high-purity, azide-free preparations for optimal and consistent activity. |
| DMSO (Anhydrous) | Solvent for preparing Amplex Red stock solution. Must be high-quality and anhydrous to prevent substrate degradation. |
| Resorufin (Sodium Salt) | Fluorescent oxidation product of Amplex Red. Used as a standard for generating a calibration curve and validating instrument performance. |
| Black-walled, Clear-bottom Microplates | Minimizes well-to-well optical crosstalk and background fluorescence while allowing for microscopic visualization if needed. |
| H₂O₂ Standard Solution | Used as a positive control to validate the assay system's responsiveness and for standard curve generation in cell-free systems. |
| Catalase | Enzyme that specifically scavenges H₂O₂. Serves as a critical negative control to confirm signal specificity. |
| Krebs-Ringer or HBSS Buffer | Physiological salt buffers for maintaining cell viability during extracellular measurement periods. |
Visualization: Amplex Red Assay Mechanism and Workflow
Diagram 1: Amplex Red Reaction and Assay Workflow
Application Note: Within a Thesis on Amplex Red Assay for Extracellular H₂O₂ Detection
Accurate quantification of extracellular hydrogen peroxide (H₂O₂) via the Amplex Red assay is foundational for research in redox signaling, oxidative stress, and drug mechanisms. This protocol details the generation of a robust standard curve and the subsequent calculation of unknown sample concentrations, critical for thesis research aiming to characterize H₂O₂ flux from cellular models or enzymatic sources.
Core Principle & Workflow
The Amplex Red/Peroxidase assay detects H₂O₂ with high sensitivity and specificity. Horseradish peroxidase (HRP) catalyzes the reaction between H₂O₂ and Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine) to produce highly fluorescent resorufin (λex/λem ~571/585 nm).
Diagram: Amplex Red to Resorufin Conversion Pathway.
Essential Research Reagent Solutions
| Reagent/Solution | Function in Assay | Critical Notes for Thesis Research |
|---|---|---|
| Amplex Red Stock (10 mM in DMSO) | Fluorogenic substrate. Stable at -20°C, protect from light. | Aliquot to avoid freeze-thaw cycles. Background oxidation can increase blanks. |
| Horseradish Peroxidase (HRP) Stock | Enzyme catalyst. Typically used at 0.2-1 U/mL final concentration. | Verify activity; source consistency is key for longitudinal thesis experiments. |
| H₂O₂ Standard Stock (e.g., 1-10 mM) | Used for generating the standard curve. Must be freshly prepared or accurately titrated. | Concentration decays. Use a molar extinction coefficient (ε₂₄₀ = 43.6 M⁻¹cm⁻¹) to verify. |
| Assay Buffer (e.g., Krebs, PBS, HBSS) | Reaction milieu. Must be phenol red-free. | Include pH control. Chelators (e.g., EDTA) may be needed to inhibit metal-catalyzed reactions. |
| Reaction Stop Solution | Optional. 1X Catalase (500 U/mL) or sodium azide (10 mM). | Halts reaction for fixed-timepoint readings; essential for non-kinetic plate readers. |
Detailed Protocol: Standard Curve Generation & Sample Analysis
A. Preparation of H₂O₂ Standard Dilution Series
- Prepare a 100 µM H₂O₂ working solution from your stock in the assay buffer.
- In a 96-well microplate, serially dilute the 100 µM solution in buffer to create standard points. A typical range is 0, 0.1, 0.2, 0.5, 1, 2, 5, 10 µM (final concentration in well).
- Run all standards in triplicate.
B. Reaction Setup for Standards and Unknowns
- Prepare the Master Reaction Mix: Combine assay buffer, Amplex Red (final 50-100 µM), and HRP (final 0.2 U/mL). Keep on ice, protected from light.
- For Standards: Add 50 µL of each H₂O₂ standard to its well. Add 50 µL of Master Reaction Mix. Total reaction volume = 100 µL.
- For Unknown Samples (e.g., cell culture supernatant): Add 50 µL of sample to the well. Add 50 µL of Master Reaction Mix. Include controls: Sample + Mix without HRP; Sample + Mix with exogenous Catalase (to confirm H₂O₂ specificity).
- Incubate at 37°C or room temperature for 30 minutes, protected from light.
- Measure fluorescence (Ex/Em ~571/585 nm).
C. Data Calculation & Quantification
- Calculate the average fluorescence value for each standard and sample replicate.
- Subtract the average fluorescence of the 0 µM standard (blank) from all other standard and sample values.
- Generate a standard curve by plotting the blank-corrected fluorescence (y-axis) against the known H₂O₂ concentration (x-axis) for the standards.
- Perform linear regression analysis. The ideal curve has an R² value >0.99.
- Use the linear equation (y = mx + c) to calculate the concentration of H₂O₂ in unknown samples.
Diagram: H₂O₂ Concentration Calculation Workflow.
Data Presentation: Standard Curve & Sample Analysis
Table 1: Representative H₂O₂ Standard Curve Data (30 min incubation)
| H₂O₂ Standard (µM) | Mean RFU (n=3) | SD | Blank-Corrected RFU |
|---|---|---|---|
| 0.0 (Blank) | 1050 | 45 | 0 |
| 0.1 | 1250 | 52 | 200 |
| 0.5 | 1850 | 61 | 800 |
| 1.0 | 2650 | 88 | 1600 |
| 2.0 | 4250 | 120 | 3200 |
| 5.0 | 9050 | 210 | 8000 |
| 10.0 | 17050 | 350 | 16000 |
Linear Regression Parameters:
- Equation: y = 1595.2x + 15.8
- R² Value: 0.9994
- Linear Range: 0.1 – 10 µM under these conditions.
Table 2: Quantification of H₂O₂ in Unknown Cell Supernatant Samples
| Sample ID & Condition | Mean RFU (n=3) | Blank-Corr. RFU (y) | Calculated [H₂O₂] (µM) | Notes |
|---|---|---|---|---|
| Ctrl Supernatant | 2200 | 1150 | 0.71 µM | Baseline extracellular [H₂O₂] |
| Drug-Treated Supernatant | 5800 | 4750 | 2.97 µM | Indicates induced H₂O₂ production |
| Drug + Catalase Control | 1080 | 30 | 0.01 µM | Confirms signal specificity to H₂O₂ |
Thesis Research Note: For extracellular H₂O₂ detection, express final sample concentrations while accounting for any sample dilution during assay setup. Normalize data to cell count or protein content as required for comparative analyses between experimental conditions. The standard curve must be generated in parallel with every experiment to control for inter-assay variability.
Solving Common Problems: Pitfalls, Optimization, and Advanced Techniques
Within the broader thesis on optimizing the Amplex Red assay for extracellular hydrogen peroxide (H₂O₂) detection, a common experimental hurdle is obtaining a low or inconsistent fluorescent signal. This Application Note systematically addresses three primary variables: cell seeding density, horseradish peroxidase (HRP) activity, and Amplex Red/Substrate limitations. Proper troubleshooting of these factors is critical for researchers, scientists, and drug development professionals employing this assay to study reactive oxygen species (ROS) in pharmacological or toxicological contexts.
The following tables summarize critical parameters and findings from current literature and experimental optimization.
Table 1: Recommended Cell Seeding Density Ranges for Common Cell Lines
| Cell Line | Recommended Density (cells/well in 96-well) | Signal Outcome at Low Density | Signal Outcome at High Density |
|---|---|---|---|
| RAW 264.7 (macrophage) | 5.0 x 10⁴ - 1.0 x 10⁵ | Low signal due to insufficient H₂O₂ production | Signal quenching, confluency-induced senescence |
| HEK 293 | 2.0 x 10⁴ - 5.0 x 10⁴ | Low basal signal | Increased background from metabolism |
| PC12 | 3.0 x 10⁴ - 7.0 x 10⁴ | Unreliable agonist response | Necrotic core, variable signal |
| Primary Neurons | 1.0 x 10⁴ - 3.0 x⁴ | Low but specific signal | High, non-specific aggregation |
Table 2: HRP Concentration and Activity Optimization
| Parameter | Typical Range | Impact on Signal | Notes |
|---|---|---|---|
| HRP Working Concentration | 0.1 - 1.0 U/mL | Signal increases with concentration up to saturation (~0.5 U/mL) | >1.0 U/mL can increase background. |
| Buffer pH (for HRP activity) | 7.4 (PBS) | Optimal at pH 7.4; activity declines sharply below pH 6.5 or above pH 8.5 | Use freshly prepared buffer. |
| Inhibitors/Interfering Substances | -- | Azide, cyanide, sulfide inhibit. Serum albumin can stabilize. | Avoid sodium azide in assay buffer. |
Table 3: Amplex Red Substrate Stability and Limitations
| Factor | Optimal Condition | Effect of Deviation |
|---|---|---|
| Amplex Red Concentration | 10 - 100 µM (50 µM standard) | Linear range up to ~100 µM; higher concentrations can self-oxidize. |
| Incubation Temperature | 37°C (cell-based); RT (enzymatic) | Increased non-enzymatic oxidation at >37°C. |
| Light Sensitivity | Protect from light | Rapid degradation, high background fluorescence. |
| Reaction Kinetics | Time-course: 30 min - 2 hr | Signal plateaus or decreases with prolonged incubation (>3 hr). |
Experimental Protocols
Protocol 1: Systematic Optimization of Cell Density
Objective: Determine the optimal cell seeding density for H₂O₂ detection in your specific cell model. Materials: Cultured cells, complete growth medium, sterile PBS, Amplex Red/HRP working solution (50 µM Amplex Red, 0.1 U/mL HRP in reaction buffer), 96-well clear-bottom black microplate, fluorescence microplate reader (λex ~540 nm, λem ~590 nm). Procedure:
- Cell Preparation: Trypsinize and count cells. Prepare serial dilutions in complete medium to cover a range (e.g., 1x10⁴ to 2x10⁵ cells/well for a 96-well plate).
- Seeding: Seed cells in triplicate for each density in 100 µL of medium per well. Include vehicle control wells (medium only, no cells). Incubate for 24 hours under normal growth conditions to allow adhesion and recovery.
- Assay Setup: Carefully remove medium and gently wash cells once with 100 µL of warm, sterile PBS.
- Reaction Incubation: Add 100 µL of Amplex Red/HRP working solution to each well. Incubate plate at 37°C, protected from light, for 60 minutes.
- Measurement: Read fluorescence immediately. Plot fluorescence intensity (RFU) vs. cell density. The optimal density is within the linear portion of the curve before plateau.
Protocol 2: Titrating HRP Activity
Objective: Establish the HRP concentration that maximizes signal-to-background ratio. Materials: Amplex Red stock (10 mM in DMSO), HRP stock (100 U/mL in reaction buffer), 30% H₂O₂ stock, reaction buffer (e.g., Krebs-Ringer phosphate buffer, pH 7.4), 96-well plate, plate reader. Procedure:
- Solution Prep: Prepare a master mix of 50 µM Amplex Red in reaction buffer. Aliquot this into a microplate.
- HRP Dilution: Create HRP dilutions in reaction buffer (e.g., 0, 0.01, 0.05, 0.1, 0.5, 1.0, 2.0 U/mL).
- Reaction: Add equal volumes of Amplex Red mix and HRP dilution to wells (final volume 100 µL). Initiate reaction by adding a low, constant concentration of H₂O₂ (e.g., 5 µM final).
- Kinetic Read: Immediately place plate in reader and take kinetic reads every minute for 10-15 minutes.
- Analysis: Calculate the initial velocity (V₀) of fluorescence increase for each HRP concentration. Plot V₀ vs. [HRP]. Select the concentration just before the plateau for subsequent assays.
Protocol 3: Assessing Substrate Integrity and Concentration
Objective: Verify that low signal is not due to substrate degradation or suboptimal concentration. Materials: Fresh and old batches of Amplex Red stock, HRP (0.1 U/mL), known standard of H₂O₂ (e.g., 10 µM), reaction buffer. Procedure:
- Standard Curve: Using fresh Amplex Red stock, prepare a standard curve of H₂O₂ (0, 1, 2, 5, 10 µM) with the Amplex Red/HRP working solution in a microplate.
- Substrate Comparison: Repeat the standard curve using the old or suspected Amplex Red stock.
- Concentration Titration: Holding H₂O₂ constant at 5 µM and HRP at 0.1 U/mL, vary the final Amplex Red concentration (1, 10, 25, 50, 100 µM).
- Incubation & Read: Incubate at 37°C for 30 min protected from light. Measure fluorescence.
- Interpretation: Compare slopes of the fresh vs. old standard curves. A shallower slope indicates substrate degradation. Determine the Amplex Red concentration where signal for 5 µM H₂O₂ saturates.
Visualizations
Title: Troubleshooting Logic Flow for Low Amplex Red Signal
Title: Amplex Red Reaction Pathway for H2O2 Detection
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Materials for Amplex Red Assay Troubleshooting
| Item | Function & Importance in Troubleshooting |
|---|---|
| Amplex Red UltraReagent (10 mM) | High-purity, stabilized substrate. Minimizes background from auto-oxidation. Essential for comparing new vs. old batches. |
| Horseradish Peroxidase (HRP), Lyophilized | Core enzyme. Reconstitute fresh for activity tests. Allows precise titration independent of substrate. |
| Hydrogen Peroxide Standard (30%, stabilized) | Provides a known stimulus for generating standard curves. Critical for quantifying HRP activity and assay linearity. |
| Cell Culture-Grade DMSO, Anhydrous | For dissolving Amplex Red stock. Ensure dryness to prevent substrate hydrolysis. |
| Krebs-Ringer Phosphate Buffer (pH 7.4) | Physiological assay buffer without interfering inhibitors (e.g., azide). Maintains HRP optimal pH. |
| Black/Clear-Bottom 96-Well Microplates | Minimize cross-talk for fluorescence reads. Clear bottom allows cell inspection/microscopy. |
| Fluorescent Microplate Reader | Equipped with ~540/590 nm filters. Kinetic reading capability is ideal for initial rate measurements. |
| Cell Counter/Hemocytometer | Accurate cell density determination is the first step in troubleshooting seeding density. |
Addressing High Background and Non-Specific Fluorescence
This Application Note provides detailed protocols for mitigating high background and non-specific fluorescence in the Amplex Red/Peroxidase assay for extracellular hydrogen peroxide (H₂O₂) detection. Within the broader thesis context, these interferences are critical confounders for accurately quantifying H₂O₂ flux from cellular systems, particularly in drug screening where subtle modulations of reactive oxygen species (ROS) are measured. The Amplex Red assay, while highly sensitive, is susceptible to artifacts from various sources, including medium components, serum, contaminating peroxidases, and auto-oxidation of the probe.
The following table summarizes common sources of non-specific fluorescence and their typical contribution to background signal, based on current literature and experimental observations.
Table 1: Primary Sources of Interference in Amplex Red Assays
| Interference Source | Typical ΔRFU (Background) | Mechanism | Conditions of High Impact |
|---|---|---|---|
| Fetal Bovine Serum (FBS) | 2000 – 5000 RFU (10% v/v) | Presence of bovine peroxidases and oxidases. | All assays with >2% serum supplementation. |
| Phenol Red in Media | 300 – 1000 RFU | Direct interaction with HRP or photo-oxidation. | Colorimetric readouts, prolonged incubation. |
| Amplex Red Auto-oxidation | 50 – 200 RFU/hr | Spontaneous, non-enzymatic oxidation to resorufin. | High pH (>8.0), light exposure, trace metals. |
| Cell Lysate Components | 500 – 2000 RFU (50 µg protein) | Endogenous peroxidases (e.g., catalase, CYP450). | Use of crude cellular extracts or freeze-thaw cycles. |
| Drug Compounds (e.g., Antioxidants) | Variable (Can quench signal) | Scavenging of H₂O₂ or direct reduction of resorufin. | High-throughput screening libraries. |
| Ambient Light Exposure | Increases baseline slope | Photo-oxidation of Amplex Red and photobleaching of resorufin. | Inadequate plate shielding during incubation. |
Optimized Protocol for Low-Background H₂O₂ Detection
Protocol 3.1: Preparation of Serum-Free, Phenol Red-Free Assay Buffer
Objective: To formulate a buffer that minimizes chemical and enzymatic background.
- Prepare a Hanks' Balanced Salt Solution (HBSS) or Krebs-Ringer Phosphate buffer, pH 7.4.
- Ensure the buffer is without phenol red and sodium bicarbonate if CO₂ control is unavailable.
- Add 0.1 mM diethylenetriaminepentaacetic acid (DTPA) to chelate trace transition metals (Fe²⁺, Cu⁺) that catalyze probe auto-oxidation.
- Filter through a 0.22 µm filter to remove particulates. Pre-warm to 37°C.
Protocol 3.2: Validation of Reagent Purity and HRP Specificity
Objective: To confirm the H₂O₂-dependence of the signal and rule out non-specific oxidation.
- Catalase Control: For every experimental condition, include two identical sample wells.
- At the assay start, add 50 U/mL Catalase (from bovine liver) to one of the duplicate wells.
- Initiate the reaction by adding the complete Amplex Red/HRP working solution to all wells.
- Measure fluorescence (Ex/Em ~571/585 nm) kinetically for 30-60 minutes.
- Data Analysis: The signal in the catalase-treated well represents non-H₂O₂-specific background. Subtract this value from the signal in the paired untreated well to obtain the H₂O₂-specific signal.
Protocol 3.3: Standard Curve Generation with Background Subtraction
Objective: To generate an accurate standard curve that accounts for matrix effects.
- Prepare a dilution series of H₂O₂ (0 to 20 µM) in the exact same buffer/media as your samples (e.g., serum-free, phenol red-free buffer).
- In a 96-well plate, add 50 µL of each H₂O₂ standard or unknown sample per well.
- Prepare Amplex Red/HRP working solution: 100 µM Amplex Red + 0.2 U/mL HRP in assay buffer. Protect from light.
- Add 50 µL of the working solution to each well. Incubate in the dark at 37°C for 30 min.
- Read fluorescence. Note: The 0 µM H₂O₂ standard (blank) defines the assay background. All sample RFU values should have the sample-specific catalase control signal (Protocol 3.2) subtracted before interpolation from the standard curve.
Visualization of Interference Pathways & Solutions
Diagram 1: Interference sources and mitigation strategies in Amplex Red assay.
Diagram 2: Optimized low-background Amplex Red assay workflow.
The Scientist's Toolkit
Table 2: Essential Reagents and Materials for Robust Amplex Red Assays
| Item | Specification/Recommended Source | Primary Function & Importance for Low Background |
|---|---|---|
| Amplex Red | High purity (>97%), lyophilized. Store desiccated at -20°C. | Probe substrate. High purity reduces pre-existing resorufin contamination. |
| Horseradish Peroxidase (HRP) | Recombinant, lyophilized, high specific activity. | Enzyme catalyst. Recombinant form avoids contaminating peroxidases found in plant extracts. |
| Catalase | From bovine liver, ≥10,000 U/mg protein. | Negative control agent. Quenches H₂O₂-specific signal; defines non-specific background. |
| DTPA | Cell culture tested, ≥99% purity. | Chelating agent. Suppresses metal-catalyzed Amplex Red auto-oxidation. |
| Assay Buffer | HBSS or Krebs-Ringer, without Phenol Red or sodium bicarbonate. | Reaction matrix. Eliminates dye-mediated interference and pH instability. |
| H₂O₂ Standard | Diluted from 30% stock, concentration verified by A240 (ε = 43.6 M⁻¹cm⁻¹). | Standard curve generation. Critical for accurate, matrix-matched quantification. |
| Optical Microplate | Black-walled, clear-bottom, tissue-culture treated. | Signal detection. Maximizes signal-to-noise, allows for kinetic reads from adherent cells. |
| Plate Reader | Fluorescence capable with temperature control (37°C). | Measurement. Kinetic reads at 37°C improve sensitivity and dynamic range. |
This application note is framed within a broader thesis research project focused on refining the Amplex Red/horseradish peroxidase (HRP) assay for the specific, sensitive, and quantitative detection of extracellular hydrogen peroxide (H₂O₂). The Amplex Red assay is a cornerstone technique in redox biology, cell signaling research, and drug development, where precise measurement of H₂O₂ production is critical. The core principle involves the HRP-catalyzed reaction of H₂O₂ with the non-fluorescent Amplex Red probe (10-acetyl-3,7-dihydroxyphenoxazine) to generate highly fluorescent resorufin. While widely adopted, the assay's sensitivity and reliability are profoundly influenced by key biochemical and kinetic parameters. This document provides optimized protocols and data-driven insights for three critical variables: probe concentration, reaction pH, and incubation time, to achieve maximum signal-to-noise ratio and robust quantitative data for extracellular H₂O₂ detection.
Table 1: Optimization of Amplex Red Probe Concentration
Conditions: 50 mM sodium phosphate buffer (pH 7.4), 0.1 U/mL HRP, 37°C, 30 min incubation, measurement of 10 µM H₂O₂ standard.
| Probe Concentration (µM) | Fluorescence Intensity (RFU) | Background Signal (RFU) | Signal-to-Noise Ratio |
|---|---|---|---|
| 5 | 12,450 | 520 | 23.9 |
| 10 | 24,800 | 980 | 25.3 |
| 20 | 48,900 | 1,550 | 31.5 |
| 50 | 49,100 | 3,900 | 12.6 |
| 100 | 49,500 | 8,200 | 6.0 |
Table 2: Optimization of Assay Buffer pH
Conditions: 20 µM Amplex Red, 0.1 U/mL HRP, 37°C, 30 min incubation, measurement of 10 µM H₂O₂ standard.
| Buffer pH | Fluorescence Intensity (RFU) | Initial Reaction Rate (RFU/min) | Assay Stability (Signal loss after 60 min, %) |
|---|---|---|---|
| 6.0 | 28,700 | 820 | 2% |
| 6.5 | 38,400 | 1,150 | 3% |
| 7.4 | 48,900 | 1,630 | 5% |
| 8.0 | 45,200 | 1,580 | 8% |
| 8.8 | 35,100 | 1,210 | 15% |
Table 3: Kinetics of Signal Development vs. Incubation Time
Conditions: 20 µM Amplex Red, 0.1 U/mL HRP, 50 mM sodium phosphate buffer (pH 7.4), 37°C, measurement of 10 µM H₂O₂ standard.
| Incubation Time (min) | Fluorescence Intensity (RFU) | Linear Regression R² Value |
|---|---|---|
| 5 | 8,150 | 0.999 |
| 10 | 16,300 | 0.999 |
| 20 | 32,600 | 0.998 |
| 30 | 48,900 | 0.995 |
| 60 | 72,100 | 0.980 |
| 90 | 80,500 | 0.920 |
Detailed Experimental Protocols
Protocol 1: Determining Optimal Amplex Red Concentration
Objective: To identify the probe concentration that maximizes the signal-to-noise ratio for H₂O₂ detection. Materials: See "The Scientist's Toolkit" below. Procedure:
- Prepare a 50 mM sodium phosphate buffer, pH 7.4.
- Prepare a 5 mM stock solution of Amplex Red in DMSO. Protect from light.
- Prepare a 10 µM H₂O₂ standard in assay buffer from a commercial stock, concentration verified by absorbance at 240 nm (ε = 43.6 M⁻¹cm⁻¹).
- In a black 96-well plate, add 50 µL of assay buffer per well.
- Add 50 µL of Amplex Red solution to final concentrations of 5, 10, 20, 50, and 100 µM (in duplicate).
- Add 50 µL of HRP solution (0.1 U/mL final concentration).
- Initiate the reaction by adding 50 µL of the 10 µM H₂O₂ standard (10 µM final) or buffer alone (for background wells).
- Incubate plate at 37°C for 30 minutes in the dark.
- Measure fluorescence (excitation/emission = 540 nm/590 nm) using a plate reader.
- Calculate Signal-to-Noise Ratio: (Mean Signal RFU - Mean Background RFU) / (Std. Dev. of Background).
Protocol 2: Optimizing Reaction pH
Objective: To determine the pH that yields maximal HRP activity and assay stability for extracellular conditions. Materials: As above, with varied buffer systems. Procedure:
- Prepare 50 mM buffers across a pH range: phosphate (pH 6.0-8.0) and Tris (pH 8.0-8.8). Verify pH with a calibrated meter.
- Prepare a master mix containing Amplex Red (20 µM final) and HRP (0.1 U/mL final) in each pH buffer.
- Dispense 150 µL of the appropriate master mix into wells of a black 96-well plate.
- Initiate reactions by adding 50 µL of 10 µM H₂O₂ standard (in the same pH buffer) to sample wells, or buffer alone to background/control wells.
- Immediately place the plate in a pre-warmed (37°C) plate reader and initiate kinetic measurements.
- Record fluorescence (Ex/Em 540/590) every minute for 60-90 minutes.
- Analyze data: a) Maximum endpoint signal at 30 min. b) Initial rate (linear slope from 0-5 min). c) Signal stability by comparing 30-min and 60-min signals.
Protocol 3: Establishing Linear Incubation Time
Objective: To define the time window during which the assay response remains linear with H₂O₂ concentration, ensuring accurate quantification. Materials: As per Protocol 1 with optimal probe concentration and pH. Procedure:
- Prepare a standard curve of H₂O₂ (e.g., 0, 2, 5, 10, 15, 20 µM) in the optimized assay buffer.
- Prepare a working solution containing Amplex Red and HRP at optimal concentrations.
- In a black 96-well plate, combine 50 µL of standard with 150 µL of the Amplex Red/HRP working solution to start the reaction.
- Incubate the plate at 37°C in the dark.
- Measure the fluorescence from the entire plate at multiple time points (e.g., 5, 10, 20, 30, 60, 90 minutes post-initiation).
- For each time point, generate a standard curve (Fluorescence vs. [H₂O₂]) and calculate the linear regression R² value.
- The optimal incubation time is the longest period that maintains an R² > 0.995 for the standard curve, ensuring robust linear quantification.
Visualization Diagrams
The Scientist's Toolkit
Table 4: Essential Research Reagent Solutions for Amplex Red Assay Optimization
| Item | Function & Rationale |
|---|---|
| Amplex Red Reagent (10-acetyl-3,7-dihydroxyphenoxazine) | The non-fluorescent substrate. Oxidized by HRP in the presence of H₂O₂ to fluorescent resorufin. Aliquot and store desiccated at ≤ -20°C, protected from light. |
| Horseradish Peroxidase (HRP), Lyophilized Powder | The enzyme catalyst. Critical for assay specificity and signal amplification. Use a high-purity, recombinant grade for consistency. Prepare fresh working solution. |
| Hydrogen Peroxide (H₂O₂) 3% Solution or Standard | The analyte and primary standard. Required for generating standard curves. Concentration must be verified spectrophotometrically for accurate quantification. |
| Dimethyl Sulfoxide (DMSO), Anhydrous | Solvent for preparing concentrated Amplex Red stock solutions (e.g., 5-20 mM). Use high-purity, sterile DMSO to prevent probe degradation. |
| Assay Buffers (Phosphate, Tris, etc.) | Maintains optimal pH for HRP activity and mimics extracellular conditions (typically pH 7.4). Must be free of azide, which inhibits HRP. |
| Black 96- or 384-Well Microplates | Plate format for fluorescence measurement. Black walls minimize optical cross-talk between wells. Use clear bottom for possible absorbance checks. |
| Fluorescent Plate Reader | Instrument for detection. Must have appropriate filters/optics for resorufin (Ex ∼540 nm / Em ∼590 nm). Kinetic capability is required for rate-based measurements. |
| Catalase (from bovine liver) | Negative control reagent. Enzymatically degrades H₂O₂. Used to confirm signal specificity (catalase should abolish signal). |
Application Notes
The accurate detection of extracellular hydrogen peroxide (H₂O₂) using the Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) assay is a cornerstone of redox biology research in contexts ranging from immune cell activation to mitochondrial function. Its principle is elegant: in the presence of horseradish peroxidase (HRP), H₂O₂ oxidizes Amplex Red to fluorescent resorufin. However, the assay's sensitivity makes it vulnerable to two major, often concurrent, confounders: exogenous peroxidase contamination and photobleaching of the fluorescent product. This document details protocols to identify, quantify, and control for these factors to ensure data integrity.
1. Quantification of Confounder Impact
Table 1: Impact and Characteristics of Key Confounders in Amplex Red Assays
| Confounder | Primary Source | Effect on Signal | Typical Magnitude of Interference | Key Diagnostic Test |
|---|---|---|---|---|
| Peroxidase Contamination | Fetal bovine serum (FBS), some cell culture media supplements, bacterial products. | False positive increase in basal & stimulated signal. | Can contribute 10-50% of total signal in untreated cellular systems. | "No-HRP" control; Heat-inactivation of serum (30 min, 56°C). |
| Photobleaching | Prolonged or intense exposure of resorufin to excitation light (~560 nm). | False negative decrease in kinetic rate and endpoint signal. | Up to 20-30% loss over 5 reads in a plate reader. Signal half-life can be <30 min under constant illumination. | Time-series control of resorufin standard in assay buffer. |
| Combined Effect | Both present in a kinetic assay. | Non-linear, time-dependent artifact obscuring true H₂O₂ kinetics. | Difficult to deconvolute without specific controls. | Parallel measurement of all controls under identical reading conditions. |
2. Detailed Experimental Protocols
Protocol A: Validating System and Reagent Purity (Peroxidase Contamination) Objective: To determine the contribution of peroxidase activity present in assay reagents or biological samples independent of added HRP. Materials: Amplex Red stock (10 mM in DMSO), Assay Buffer (e.g., Krebs-Ringer phosphate, pH 7.4), Biological sample (e.g., cell culture supernatant, serum-containing media), H₂O₂ standard, microplate reader capable of fluorescence detection (λex/~560 nm, λem/~590 nm). Procedure:
- Prepare a master mix of Assay Buffer containing Amplex Red (final 50-100 µM).
- Aliquot the master mix into wells of a 96-well plate.
- For Test Wells, add biological sample. For Control Wells, add an equal volume of buffer or heat-inactivated sample.
- CRITICAL: Do not add HRP to any well in this protocol.
- Initiate kinetic fluorescence measurement immediately, reading every 5 minutes for 60-90 minutes.
- A steady increase in fluorescence in Test Wells versus flat Control Wells indicates significant peroxidase contamination.
Protocol B: Characterizing and Mitigating Photobleaching Objective: To quantify the rate of photobleaching under your specific instrument settings and establish a correction protocol. Materials: Resorufin standard (or fully reacted Amplex Red/H₂O₂/HRP mixture), Assay Buffer, opaque-walled 96-well plate. Procedure:
- Prepare a solution of resorufin in Assay Buffer at a fluorescence intensity approximating the maximum of your experiment (e.g., 1-10 µM).
- Aliquot this solution into all wells of a plate column (n≥6).
- Program the plate reader for your intended experiment (kinetic reads every X minutes for Y duration, with shaking before reads if used).
- Run the protocol. The observed decay in fluorescence over time is the photobleaching rate.
- Mitigation Strategy: Optimize instrument settings: use the lowest acceptable gain/detector voltage, reduce measurement time per well, employ a neutral density filter if available, and minimize the number of read cycles. Data can be corrected post-hoc using the decay constant derived from this control.
Protocol C: Integrated Assay with Confounder Controls Objective: To run an Amplex Red assay for cellular H₂O₂ release with necessary controls for data correction. Workflow:
- Seed cells in a 96-well plate. Include blank wells (no cells) for all control types.
- On the day of the assay, prepare fresh Amplex Red/HRP working solution in validated, low-background buffer.
- For each experimental condition (e.g., drug treatment), set up the following quadrupicate wells:
- Experimental Well: Cells + Amplex Red/HRP working solution.
- No-HRP Control: Cells + Amplex Red working solution without HRP.
- No-Cell Control: Amplex Red/HRP working solution only (defines assay background).
- Photobleaching Control: A well containing a fixed concentration of resorufin or a high H₂O₂ standard.
- After adding treatments/work solution, immediately place the plate in the pre-heated (37°C) reader.
- Run the kinetic protocol. The corrected fluorescence for each experimental well can be approximated as:
Fluorescence(Experimental) - Fluorescence(No-HRP Control). Compare all values to the photobleaching control trace.
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions for Controlled Amplex Red Assays
| Item | Function & Rationale |
|---|---|
| Amplex Red (High-Purity, Lyophilized) | Probe substrate. Use lyophilized form from aliquoted, desiccated stocks to prevent degradation. |
| HRP, Recombinant or Highly Purified | Reaction enzyme. Use a consistent, specific lot with defined activity to standardize kinetics. |
| Resorufin Sodium Salt | Critical standard for generating calibration curves and quantifying photobleaching rates. |
| Charcoal-Stripped or Dialyzed FBS | Serum supplement with low molecular weight components (like peroxidases) removed to reduce contamination. |
| Horseradish Peroxidase Inhibitor (e.g., Sodium Azide) | Used diagnostically to confirm HRP-dependent signal in follow-up experiments. CAUTION: Toxic. |
| Opaque-Walled or Black Microplates | Minimizes cross-talk and reduces the potential for ambient light-induced photobleaching. |
| Krebs-Ringer Phosphate Buffer (pH 7.4) | A well-defined, low-fluorescence physiological buffer; preferable over phenol red-containing media for reading. |
Visualizations
Diagram 1: Core Assay and Confounder Interference Pathways
Diagram 2: Experimental Workflow for Confounder Control
Application Notes
Within a broader thesis on optimizing the Amplex Red assay for extracellular hydrogen peroxide (H₂O₂) detection, integrating multiplexed endpoints with stopped-flow kinetics presents a powerful strategy for high-content mechanistic analysis. This approach allows researchers to deconvolute the complex interplay between reactive oxygen species (ROS) flux, cellular health, and real-time enzymatic kinetics, which is critical in drug development for oncology, neurodegeneration, and inflammatory diseases.
Key Advantages:
- Contextual Kinetics: Stopped-flow measurements provide precise, millisecond-resolution data on H₂O₂ generation or scavenging rates by isolated enzymes (e.g., NADPH oxidases, catalase). Multiplexing these with endpoint cell viability assays (e.g., MTT, Calcein-AM) in parallel or sequential experiments clarifies whether altered H₂O₂ kinetics directly correlate with cytotoxicity or cytoprotection.
- Artifact Mitigation: The Amplex Red assay can be influenced by compound autofluorescence, peroxidase activity, or changes in pH. Using a stopped-flow system minimizes these interferences during kinetic reads, while a separate, multiplexed viability check confirms that observed kinetic changes are not secondary to massive cell death.
- High-Throughput Screening (HTS) Triage: This combined strategy efficiently identifies lead compounds. Initial stopped-flow screens can identify potent modulators of H₂O₂ kinetics from pure enzyme systems. Subsequent multiplexed assays in live cells validate target engagement and immediately flag compounds with overt toxicity.
Quantitative Data Summary: The following tables consolidate key metrics from representative studies employing these advanced strategies.
Table 1: Comparison of Stopped-Flow vs. Conventional Plate Reader for Amplex Red Kinetics
| Parameter | Stopped-Flow Spectrofluorometer | Conventional Microplate Reader | Advantage of Stopped-Flow |
|---|---|---|---|
| Dead Time | < 5 ms | 1-10 seconds | Captures rapid initial rates |
| Mixing Efficiency | Highly efficient, turbulent | Laminar, diffusion-limited | Eliminates lag artifacts |
| Sample Volume | 50-200 µL per shot | 100-300 µL per well | Conserves precious enzymes/compounds |
| Data Point Density | 1000+ points per second | 1-2 points per minute | Enables robust kinetic modeling |
| Typical ( K_m) App for HRP | ~20 µM | ~35-50 µM | More accurate determination |
Table 2: Impact of Multiplexing Amplex Red with Viability Assays in Cell-Based Studies
| Cell Line / Stimulus | Amplex Red Signal (H₂O₂) | Viability Assay (MTT) | Interpretation | Reference Class |
|---|---|---|---|---|
| A549 cells, Antimycin A | ↑ 250% | ↓ 60% | Increased ROS production correlates with cytotoxicity. | [Model Toxicol.] |
| RAW 264.7 cells, LPS | ↑ 400% | 95% | Inflammatory burst is independent of acute toxicity. | [Model Inflamm.] |
| HepG2 cells, Nrf2 Activator | ↓ 40% | ↑ 120% | Antioxidant response enhances cell proliferation. | [Cytoprotection] |
| SH-SY5Y cells, Aβ(1-42) | ↑ 180% | ↓ 45% | Amyloid-β induced oxidative stress is neurotoxic. | [Neurodegeneration] |
Experimental Protocols
Protocol 1: Stopped-Flow Kinetics of H₂O₂ Scavenging by Catalase Using Amplex Red
Objective: To determine the kinetic parameters of catalase using the Amplex Red/HRP system in a stopped-flow spectrofluorometer.
Research Reagent Solutions:
| Item | Function |
|---|---|
| Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) | Fluorogenic probe, reacts with H₂O₂ in presence of HRP to yield fluorescent resorufin. |
| Horseradish Peroxidase (HRP) | Enzyme that catalyzes the reaction between Amplex Red and H₂O₂. |
| Catalase (from bovine liver) | Target enzyme that scavenges H₂O₂, kinetics of which are being measured. |
| Hydrogen Peroxide (H₂O₂) Stock | Substrate for both catalase and the Amplex Red/HRP system. |
| Potassium Phosphate Buffer (50 mM, pH 7.4) | Physiological pH buffer for all reactions. |
| Stopped-Flow Spectrofluorometer | Instrument for rapid mixing and ultra-fast fluorescence measurement. |
Methodology:
- Prepare Solutions:
- Syringe A: Amplex Red (20 µM) and HRP (0.1 U/mL) in potassium phosphate buffer.
- Syringe B: A range of H₂O₂ concentrations (e.g., 10, 25, 50, 100, 200 µM) with a fixed concentration of catalase (e.g., 10 nM) in potassium phosphate buffer.
- Instrument Setup:
- Equilibrate stopped-flow instrument to 25°C.
- Set excitation to 560 nm (slit 2 nm) and emission to 590 nm (slit 10 nm) using a cut-off filter (>570 nm).
- Set a push volume to achieve a 1:1 mixing ratio and a total observation time of 2-5 seconds.
- Kinetic Measurement:
- Load Syringe A and Syringe B.
- Initiate rapid mixing and data acquisition. Perform a minimum of 5 shots per H₂O₂ concentration and average the traces.
- The observed fluorescence increase corresponds to H₂O₂ not scavenged by catalase during the reaction delay time.
- Data Analysis:
- Fit the initial linear portion (first 5-10%) of each averaged fluorescence trace to obtain the initial velocity (V₀) of resorufin formation.
- Plot V₀ vs. [H₂O₂]. Fit data to the Michaelis-Menten equation to derive apparent ( Km ) and ( V{max} ) for catalase under these competitive conditions.
Protocol 2: Multiplexed Amplex Red and MTT Viability Assay in Adherent Cells
Objective: To simultaneously measure stimulus-induced extracellular H₂O₂ production and its impact on cell viability in a 96-well format.
Research Reagent Solutions:
| Item | Function |
|---|---|
| Amplex Red/HRP Working Solution | Contains Amplex Red (50 µM) and HRP (0.1 U/mL) in HBSS or phenol-red-free culture medium. |
| MTT Reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) | Yellow tetrazole, reduced to purple formazan by metabolically active cells. |
| Cell Lysis/Solubilization Solution | Typically DMSO or SDS-based buffer, dissolves formazan crystals. |
| HBSS (Hanks' Balanced Salt Solution) | Salt and glucose solution for maintaining cells during assay, without phenol red. |
| Test Compound(s) & Stimuli/Inhibitors | Pharmacological agents being studied for their effect on ROS and viability. |
Methodology:
- Cell Seeding & Treatment:
- Seed cells in a flat, clear-bottom 96-well plate at optimal density (e.g., 10⁴ cells/well). Culture for 24 hours.
- Treat cells with test compounds, stimulants (e.g., PMA, TNF-α), or inhibitors for the desired time.
- Amplex Red Incubation (H₂O₂ Detection):
- Carefully aspirate culture medium and wash cells once with warm HBSS.
- Add 100 µL of Amplex Red/HRP working solution to each well.
- Immediately place plate in a pre-warmed (37°C) microplate reader.
- Measure fluorescence (Ex/Em ~560/590 nm) kinetically every 5 minutes for 60-90 minutes.
- MTT Assay (Viability Measurement):
- After the final Amplex Red read, carefully aspirate the Amplex Red solution.
- Add 100 µL of fresh culture medium containing 0.5 mg/mL MTT to each well.
- Incubate for 2-4 hours at 37°C.
- Carefully aspirate the medium without disturbing the formed formazan crystals.
- Add 100 µL of DMSO to solubilize the crystals. Shake gently for 10 minutes.
- Measure absorbance at 570 nm (reference ~650 nm) in the plate reader.
- Data Analysis:
- H₂O₂: Calculate the slope (RFU/min) from the linear phase of the Amplex Red fluorescence curve for each well.
- Viability: Normalize MTT absorbance values to vehicle control wells (set as 100%).
- Correlate normalized H₂O₂ production rates with normalized cell viability for each treatment condition.
Mandatory Visualization
Diagram 1: Multiplexed assay workflow for ROS & viability
Diagram 2: Amplex Red H₂O₂ detection reaction pathway
Diagram 3: Decision logic for compound triage via multiplexing
Validation and Comparison: How Amplex Red Stacks Up Against Other H₂O₂ Detection Methods
1. Introduction & Thesis Context Within a broader thesis investigating extracellular hydrogen peroxide (H₂O₂) dynamics using the Amplex Red assay, rigorous validation is paramount. The Amplex Red/HRP system, while highly sensitive, can produce artifacts from non-specific peroxidase activity or non-enzymatic oxidation. This document details application notes and protocols employing positive/negative controls and enzymatic specificity, using catalase, to validate that the measured signal truly originates from extracellular H₂O₂.
2. Research Reagent Solutions Toolkit Table 1: Essential Reagents for Validation Experiments
| Reagent/Solution | Function in Validation | Key Consideration |
|---|---|---|
| Amplex Red Reagent | Fluorescent probe oxidized in the presence of H₂O₂ and HRP to generate resorufin. | Sensitivity to photobleaching; prepare fresh or from frozen aliquots. |
| Horseradish Peroxidase (HRP) | Enzyme that catalyzes the oxidation of Amplex Red by H₂O₂. | Source and purity can affect background signal. |
| Catalase (from bovine liver) | Specificity Control Enzyme. Catalyzes the decomposition of H₂O₂ to H₂O and O₂. Used to confirm the signal source. | High specific activity (>2000 U/mg). Heat-inactivated control is crucial. |
| Exogenous H₂O₂ Standard | Positive Control. Provides a known signal for assay calibration and system functionality check. | Must be calibrated spectrophotometrically (ε240 = 43.6 M⁻¹cm⁻¹). |
| Superoxide Dismutase (SOD) | Specificity Control. Scavenges superoxide (O₂˙⁻), which can indirectly generate H₂O₂ or cause artifacts. | Used to rule out signal contribution from O₂˙⁻. |
| Heat-Inactivated Catalase | Negative Control for Catalase Specificity. Confirms that observed inhibition is enzymatic, not artifactual. | Prepare by heating catalase at 95°C for 15-30 minutes. |
| Cell Culture Medium (Phenol Red-free) | Assay medium. Phenol Red can interfere with fluorescence measurements. | Should be pre-warmed and pH-adjusted. |
| Specific Agonist/Inhibitor | Pharmacological tool to stimulate or inhibit cellular H₂O₂ production (e.g., PMA for NADPH oxidase). | Validates the biological relevance of the detected signal. |
3. Core Validation Protocols
3.1. Protocol A: Establishing the Assay Linear Range & Positive Control Objective: To verify the performance of the Amplex Red/HRP system and define the linear range for H₂O₂ quantification. Procedure:
- Prepare a reaction buffer (e.g., Krebs-Ringer buffer, pH 7.4).
- Add Amplex Red (final conc. 50 µM) and HRP (final conc. 0.1 U/mL) to the buffer.
- In a 96-well plate, add 100 µL of the Amplex Red/HRP mix per well.
- Spike in known concentrations of exogenous H₂O₂ standard (0, 0.5, 1, 2, 5, 10 µM) in triplicate.
- Incubate at 37°C for 30 minutes protected from light.
- Measure fluorescence (Ex/Em ≈ 560/590 nm).
- Plot fluorescence vs. [H₂O₂] to generate a standard curve and determine the linear range (R² > 0.98). Data Output: See Table 2, Standard Curve Data.
3.2. Protocol B: Specificity Validation using Catalase Objective: To confirm that the fluorescent signal in biological samples is specifically derived from H₂O₂. Procedure:
- Plate and treat cells according to the experimental design of the thesis.
- Prior to assay, pre-treat sample aliquots (or wells) with one of the following for 15-30 minutes: a. Active Catalase (250-1000 U/mL final). b. Heat-Inactivated Catalase (same protein concentration as active). c. Buffer only (untreated control).
- Perform the Amplex Red assay (as per thesis methodology) on all conditions.
- Compare signal reduction. A significant reduction (>70-90%) with active catalase, but not with heat-inactivated catalase, confirms H₂O₂-specific signal. Data Output: See Table 3, Catalase Specificity Test.
3.3. Protocol C: Integrated Workflow for Validated Extracellular H₂O₂ Measurement Objective: A step-by-step protocol for a validated sample measurement within the thesis framework.
- Pre-assay: Generate a H₂O₂ standard curve (Protocol A) on the same plate as samples.
- Sample Preparation: Collect extracellular medium from stimulated cells (e.g., treated with PMA). Centrifuge to remove any detached cells.
- Specificity Test Setup: Aliquot sample medium into three separate reaction wells:
- Well 1: Sample + Amplex Red/HRP (Total Signal).
- Well 2: Sample + Amplex Red/HRP + Active Catalase (1000 U/mL) (Specificity Control).
- Well 3: Sample + Amplex Red/HRP + Heat-Inactivated Catalase (Negative Control).
- Incubation: Incubate plate at 37°C for 30-60 min, protected from light.
- Detection: Measure fluorescence.
- Calculation: Subtract the signal from the Active Catalase well from the Total Signal to determine the catalase-sensitive (i.e., H₂O₂-specific) signal. Use the standard curve for quantification.
4. Data Presentation
Table 2: Standard Curve Data for Amplex Red Assay (Positive Control)
| Hydrogen Peroxide (µM) | Fluorescence (RFU, Mean ± SD, n=3) | Linearity Check |
|---|---|---|
| 0.0 | 1250 ± 150 | Baseline |
| 0.5 | 4350 ± 320 | Linear |
| 1.0 | 7450 ± 410 | Linear |
| 2.0 | 13800 ± 780 | Linear |
| 5.0 | 32000 ± 2100 | Linear |
| 10.0 | 58500 ± 3500 | Start of plateau |
Table 3: Catalase Specificity Test in PMA-Stimulated Leukocytes
| Sample Condition | Fluorescence (RFU, Mean ± SD, n=4) | % of Total Signal | Interpretation |
|---|---|---|---|
| PMA-Stimulated (Total Signal) | 25,000 ± 1,800 | 100% | Reference maximum signal. |
| + Active Catalase | 3,200 ± 450 | 12.8% | Catalase-sensitive signal (87.2%) is H₂O₂-specific. |
| + Heat-Inact. Catalase | 24,100 ± 1,650 | 96.4% | Confirms inhibition is enzymatic. |
| Unstimulated Cells | 4,100 ± 600 | 16.4% | Baseline extracellular H₂O₂. |
5. Diagrams
Diagram 1: H2O2 Detection and Validation Mechanism (94 chars)
Diagram 2: Step-by-Step Validation Protocol (92 chars)
This application note supports a thesis investigating the Amplex Red assay as a gold standard for specific, extracellular hydrogen peroxide (H₂O₂) detection. A critical component of validating this thesis is a direct comparison with the commonly used, yet often misapplied, fluorescent probe DCFDA (also known as DCFH-DA). While Amplex Red is explicitly designed for extracellular H₂O₂, DCFDA is primarily a cell-permeable probe for intracellular reactive oxygen species (ROS). This analysis delineates their distinct mechanisms, applications, and pitfalls to guide researchers in selecting the appropriate tool for extracellular oxidant measurement in drug discovery and mechanistic studies.
Mechanism and Specificity Comparison
Amplex Red: In the presence of horseradish peroxidase (HRP), Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) reacts specifically with H₂O₂ in a 1:1 stoichiometry to produce highly fluorescent resorufin (Ex/Em ~571/585 nm). The assay is conducted extracellularly; the probe and HRP are not cell-permeable, ensuring detection of only secreted H₂O₂.
DCFDA (2',7'-Dichlorofluorescin diacetate): This cell-permeable dye is deacetylated by intracellular esterases to non-fluorescent DCFH, which is trapped inside cells. DCFH is oxidized by a broad range of intracellular ROS (e.g., •OH, ONOO⁻, and non-specifically by H₂O₂ via cellular peroxidases) to fluorescent DCF (Ex/Em ~495/529 nm). Its use for extracellular detection is inappropriate without careful experimental modification and validation.
The fundamental signaling pathways and assay contexts are distinct, as summarized in the following diagram.
Quantitative Comparison Table
Table 1: Direct Comparison of Key Assay Parameters
| Parameter | Amplex Red Assay | DCFDA/DCFH-DA Assay |
|---|---|---|
| Primary Target | Extracellular H₂O₂ | Broad intracellular ROS |
| Specificity | High for H₂O₂ | Low; multiple oxidants |
| Stoichiometry | 1:1 (H₂O₂:Resorufin) | Non-stoichiometric, variable |
| Typical Dynamic Range | ~0.1 to 10 µM H₂O₂ | Not quantifiable for [H₂O₂] |
| Key Enzyme | Exogenous Horseradish Peroxidase (HRP) | Intracellular esterases & peroxidases |
| Signal Localization | Extracellular medium | Intracellular |
| Common Artifacts | Phenol red interference, HRP inhibitors | Auto-oxidation, photo-oxidation, cytotoxicity |
| Quantitative Potential | High (can use standard curve) | Low to moderate (semi-quantitative) |
| Best Suited For | Kinetic measurement of H₂O₂ production/release from cells, enzymes, or drugs. | Detecting general shifts in intracellular oxidative stress. |
Detailed Experimental Protocols
Protocol 1: Extracellular H₂O₂ Detection Using Amplex Red
This protocol is central to the thesis for validating drug effects on extracellular H₂O₂ flux.
I. Materials (The Scientist's Toolkit) Table 2: Essential Reagents for Amplex Red Assay
| Item | Function & Notes |
|---|---|
| Amplex Red Reagent | Probe substrate (10-acetyl-3,7-dihydroxyphenoxazine). Prepare stock in DMSO, store at -20°C protected from light. |
| Horseradish Peroxidase (HRP) | Enzyme catalyst. Use high-purity, lyophilized powder. Reconstitute in assay buffer. |
| H₂O₂ Standard (e.g., 30% w/w) | For generating a standard curve. Standardize concentration spectrophotometrically (A₂₄₀, ε=43.6 M⁻¹cm⁻¹). |
| Hanks' Balanced Salt Solution (HBSS) or Phenol-red-free buffer | Assay buffer. Phenol red must be omitted as it absorbs/fluoresces at similar wavelengths. |
| 96-well Black Microplate | Optically clear bottom for fluorescence readings. |
| Fluorescence Microplate Reader | Equipped with filters/optics for ~571/585 nm (Ex/Em). |
II. Workflow
III. Step-by-Step Method
- Standard Curve: Prepare serial dilutions of H₂O₂ (e.g., 0, 0.5, 1, 2, 5, 10 µM) in assay buffer.
- Working Solution: Prepare Amplex Red/HRP working solution in phenol-red-free buffer immediately before use. Final recommended concentrations: 50 µM Amplex Red, 0.1 U/mL HRP.
- Assay Setup: In a black 96-well plate, add 50 µL of sample (cell culture supernatant, buffer control, or H₂O₂ standard) to 50 µL of the working solution. Run in triplicate.
- Incubation: Cover plate, incubate at 37°C (or desired temperature) for 30-60 minutes protected from light.
- Measurement: Read fluorescence using a microplate reader (Ex/Em ~571/585 nm).
- Analysis: Subtract the fluorescence of the no-H₂O₂ control (blank) from all values. Plot standard curve (Fluorescence vs. [H₂O₂]) and calculate sample H₂O₂ concentration.
Protocol 2: Intracellular ROS Detection Using DCFDA (for Contextual Comparison)
This protocol highlights the contrasting methodology for intracellular assessment.
I. Materials (Key Items)
| Item | Function & Notes |
|---|---|
| DCFDA (DCFH-DA) | Cell-permeable probe. Prepare stock in DMSO, store at -20°C protected from light. |
| Cell Culture Medium | Phenol-red-free, serum-free for loading (serum contains esterases). |
| Positive Control (e.g., tert-Butyl hydroperoxide) | Inducer of oxidative stress to validate assay response. |
| Fluorescence Microplate Reader | Filters for ~495/529 nm (Ex/Em). |
II. Step-by-Step Method
- Cell Preparation: Seed cells in a black 96-well plate with clear bottom. Grow to desired confluence.
- Probe Loading: Wash cells with PBS. Load cells with 10-20 µM DCFDA in phenol-red-free, serum-free medium for 30-45 minutes at 37°C.
- Wash: Thoroughly wash cells 2-3 times with warm buffer to remove extracellular probe.
- Treatment & Measurement: Add fresh medium containing test compounds or vehicle. Immediately place plate in a pre-warmed (37°C) plate reader. Measure fluorescence (Ex/Em ~495/529 nm) kinetically (e.g., every 5-15 min for 1-2 hours). Note: This protocol measures intracellular oxidation.
- Extracellular Modification (Advanced): To attempt extracellular detection, one can load and trap DCFH in cells, then lyse them and add the lysate to an extracellular reaction mix. This is complex and not recommended for routine extracellular H₂O₂ measurement.
For the specific thesis focus on extracellular H₂O₂—a key signaling molecule and drug target in pathologies like inflammation and cancer—the Amplex Red assay is the unequivocally superior and appropriate choice. It provides specific, quantitative, kinetic data on secreted H₂O₂. DCFDA is unsuitable for this purpose; its application should be restricted to reporting gross changes in intracellular oxidative stress. Misapplication of DCFDA for extracellular H₂O₂ detection risks generating misleading data in drug development pipelines. Validating drug mechanisms requires the specificity offered by the Amplex Red/HRP system.
Strengths and Weaknesses Relative to Luminol-Based Chemiluminescence and Electrochemical Sensors
This application note supports a thesis investigating the Amplex Red/Peroxidase assay for specific, quantitative detection of extracellular hydrogen peroxide (H₂O₂) in biological systems. A critical evaluation of alternative prevalent methodologies—luminol-based chemiluminescence and electrochemical sensors—is essential for justifying methodological choices, optimizing experimental design, and accurately interpreting data. This document provides a comparative analysis, structured protocols, and key resources for researchers.
Comparative Analysis: Amplex Red vs. Luminol vs. Electrochemical Sensors
Core Principles and Mechanisms
- Amplex Red Assay: A fluorogenic probe. In the presence of horseradish peroxidase (HRP), H₂O₂ oxidizes Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) to resorufin, a highly fluorescent product (Ex/Em ~571/585 nm).
- Luminol-Based Chemiluminescence: Luminol (5-amino-2,3-dihydro-1,4-phthalazinedione) is oxidized by H₂O₂ under catalytic conditions (e.g., HRP, ferricyanide), producing an excited-state product that emits light (~425 nm) upon relaxation.
- Electrochemical Sensors: Typically employ an electrode modified with enzymes (e.g., horseradish peroxidase) or selective catalysts. H₂O₂ reduction or oxidation at the electrode surface generates a measurable current proportional to concentration.
Quantitative Comparison Table
Table 1: Performance Characteristics of H₂O₂ Detection Methods
| Feature | Amplex Red / HRP Assay | Luminol-Based Chemiluminescence | Electrochemical (Enzymatic) Sensor |
|---|---|---|---|
| Detection Principle | Fluorogenic | Chemiluminescent | Amperometric/Potentiometric |
| Sensitivity (Typical LOD) | ~10-100 nM | ~1-10 nM (can be higher) | ~10-100 nM |
| Dynamic Range | Up to ~100 µM | Several orders of magnitude | Linear over a wide range (µM-mM) |
| Specificity for H₂O₂ | High (with HRP) | Low (reacts with ROS, ONOO⁻) | High (with HRP or selective catalyst) |
| Temporal Resolution | Seconds to minutes (plate reading) | Milliseconds to seconds (real-time capable) | Milliseconds (real-time, continuous) |
| Spatial Mapping | Possible with microscopy | Difficult, low resolution | Excellent (microelectrodes) |
| Throughput | High (plate reader) | Medium to High | Low (single-point) to Medium (array) |
| Sample Consumption | Low to Medium | Very Low | Very Low |
| Key Interferences | Cellular reductants (e.g., ascorbate), HRP inhibitors | Other oxidants (e.g., hypochlorite), metal ions, pH | Electroactive species (ascorbate, urate), electrode fouling |
| Ease of Use | Simple, endpoint or kinetic | Requires careful reagent optimization & injector | Requires equipment & calibration expertise |
| Cost per Sample | Low | Low | High (initial setup, electrode maintenance) |
Table 2: Suitability for Common Research Applications
| Application Context | Recommended Method | Rationale |
|---|---|---|
| High-throughput screening of drug effects on H₂O₂ production | Amplex Red | Robust, plate-reader compatible, quantitative. |
| Real-time kinetics of rapid H₂O₂ bursts (e.g., NADPH oxidase activity) | Luminol or Electrochemical | Superior temporal resolution. |
| In vivo or intravital H₂O₂ monitoring | Electrochemical | Only method allowing continuous, real-time measurement in live tissue. |
| Mapping H₂O₂ gradients (e.g., wound healing, root tips) | Electrochemical | Spatial resolution with scanning probes. |
| Detection in complex, metal-rich media (e.g., blood) | Amplex Red (with caution) | Less prone to metal-catalyzed non-specific signal than luminol. |
| Specific detection in systems with multiple ROS | Amplex Red or Electrochemical | Higher specificity than luminol. |
Detailed Experimental Protocols
Protocol 1: Amplex Red Assay for Extracellular H₂O₂ in Cell Culture
Purpose: To quantify steady-state or stimulated extracellular H₂O₂ release from adherent cells. Key Reagents & Solutions: See "The Scientist's Toolkit" (Section 5). Workflow:
- Cell Preparation: Plate cells in a clear-bottomed, black-walled 96-well plate. Grow to desired confluence. Serum-starve if required by experiment.
- Reagent Preparation: Prepare 1X Reaction Buffer (KRPH or HBSS, pH 7.4). Freshly prepare Amplex Red Working Solution: 50 µM Amplex Red, 0.1 U/mL HRP in 1X Reaction Buffer. Protect from light.
- Assay Execution: a. Remove cell culture medium and gently wash cells with warm 1X Reaction Buffer (2 x 100 µL). b. Add 90 µL of Amplex Red Working Solution per well. c. Initiate the reaction by adding 10 µL of stimulus (e.g., growth factor, drug) or vehicle control directly to the well. Mix gently by orbital shaking. d. Immediately place plate in a pre-warmed (37°C) fluorescence microplate reader.
- Data Acquisition: Measure fluorescence (Ex 530-560 nm / Em 580-600 nm, e.g., Ex 560/Em 590) kinetically every 1-5 minutes for 30-120 minutes.
- Quantification: Generate a standard curve (0-100 µM H₂O₂) in wells without cells. Subtract the fluorescence of a no-HRP control. Calculate H₂O₂ concentration from the standard curve slope.
Amplex Red Assay Workflow for Cell-Based H₂O₂ Detection
Protocol 2: Luminol-Based Chemiluminescence for Real-Time H₂O₂ Detection
Purpose: To capture rapid, real-time dynamics of H₂O₂ production in a cell-free enzymatic system (e.g., purified enzyme kinetics). Key Reagents & Solutions: See "The Scientist's Toolkit" (Section 5). Workflow:
- Instrument Setup: Pre-warm a luminometer with injector to 37°C. Set to measure in continuous mode with 0.1-1 second intervals.
- Reagent Preparation: Prepare Luminol Working Solution (e.g., 100 µM luminol, 10 µg/mL HRP in assay buffer). Protect from light. Prepare H₂O₂ standard or enzyme/substrate solution.
- Assay Execution: a. Pipette 90 µL of Luminol Working Solution into a luminometer tube or white plate well. b. Place in luminometer and initiate background reading for 10-30 seconds. c. Use the injector to rapidly add 10 µL of the H₂O₂-containing sample or stimulus. d. Record luminescence for 2-10 minutes.
- Data Analysis: Plot relative light units (RLU) vs. time. Quantify by integrating the area under the curve (AUC) or measuring peak height.
Luminol Chemiluminescence Real-Time Assay Workflow
Signaling Pathway Contextualization
Extracellular H₂O₂ Production and Detection in Redox Signaling
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for H₂O₂ Detection Research
| Item / Reagent | Function & Importance | Example Supplier / Cat. # (Representative) |
|---|---|---|
| Amplex Red Reagent | Fluorogenic substrate. Specific oxidation by H₂O₂:HRP yields fluorescent resorufin. | Thermo Fisher Scientific, A12222 |
| Horseradish Peroxidase (HRP) | Essential enzyme catalyst for both Amplex Red and luminol assays. Purity is critical. | Sigma-Aldrich, P8375 |
| Luminol (sodium salt) | Chemiluminescent substrate. Oxidation yields light emission, enabling high sensitivity. | Sigma-Aldrich, 123072 |
| Hydrogen Peroxide, 30% (w/w) | Primary standard for generating calibration curves. Must be diluted fresh and quantified. | Sigma-Aldrich, H1009 |
| Catalase (from bovine liver) | Negative control enzyme. Scavenges H₂O₂; confirms signal specificity. | Sigma-Aldrich, C1345 |
| Krebs-Ringer Phosphate HEPES (KRPH) Buffer | Common physiological assay buffer for extracellular measurements, minimizes artifacts. | MilliporeSigma, K4002 (or prepared in-lab) |
| Clear-Bottom Black-Wall 96-Well Plate | Optimal for fluorescence assays (minimizes cross-talk). | Corning, 3904 |
| White Opaque 96-Well Plate | Optimal for luminescence assays (maximizes light capture). | Corning, 3912 |
| H₂O₂ Electrochemical Sensor | Microsensor for real-time, spatially resolved detection. Requires specific potentiostat. | Unisense, H₂O₂-100 |
| Potentiostat / Galvanostat | Instrument required to operate and read electrochemical sensors. | Palmsens, EmStat4S |
Correlating Extracellular H₂O₂ with Intracellular ROS Probes for a Complete Picture
Within the broader thesis on the Amplex Red assay for extracellular hydrogen peroxide (H₂O₂) detection, a critical gap exists in correlating these extracellular measurements with intracellular reactive oxygen species (ROS) dynamics. This application note provides integrated protocols to simultaneously quantify extracellular H₂O₂ release and intracellular ROS production, enabling a comprehensive redox profile of biological systems relevant to pharmacology, toxicology, and cell signaling research.
Key Research Reagent Solutions
The following table details essential materials for performing these correlated assays.
| Item | Function & Rationale |
|---|---|
| Amplex Red (10-Acetyl-3,7-dihydroxyphenoxazine) | A cell-impermeable probe that, in the presence of HRP, reacts stoichiometrically with extracellular H₂O₂ to form fluorescent resorufin (Ex/Em ~571/585 nm). |
| Horseradish Peroxidase (HRP) | Enzyme required to catalyze the reaction between Amplex Red and H₂O₂. Always used in excess. |
| Cell-Permeable Intracellular ROS Probes (e.g., H2DCFDA, DHE) | H2DCFDA is oxidized by various intracellular ROS to fluorescent DCF. Dihydroethidium (DHE) is oxidized specifically by superoxide to form 2-hydroxyethidium. |
| Catalase | Negative control enzyme that scavenges H₂O₂. Confirms Amplex Red signal specificity. |
| Superoxide Dismutase (SOD) | Converts superoxide to H₂O₂. Can be used to modulate signals in both assays. |
| Specific ROS Inducers/Inhibitors (e.g., Antimycin A, PMA, N-Acetylcysteine, Apocynin) | Pharmacological tools to manipulate ROS production and validate probe responses. |
| Fluorescence Microplate Reader | Instrument capable of kinetic reads and multiple wavelength pairs (e.g., 571/585 nm for Amplex Red, 488/525 nm for DCF, 518/605 nm for Ethidium/2-OH-E+). |
Integrated Experimental Protocols
Protocol 1: Concurrent Measurement in a Microplate Format
This protocol is optimized for adherent cells in a 96-well plate, allowing parallel kinetic tracking of both extracellular and intracellular ROS.
Materials:
- HBSS or Phenol Red-free culture buffer, pH 7.4
- Amplex Red stock solution (10 mM in DMSO)
- HRP stock (100 U/mL in buffer)
- H2DCFDA stock (10 mM in DMSO)
- Test compounds, stimulants, or inhibitors
Procedure:
- Cell Preparation: Seed cells in a clear-bottom black-walled 96-well plate. Grow to desired confluence (typically 80-90%). Serum-starve if required by the experimental model.
- Probe Loading: Prepare a working solution of H2DCFDA in pre-warmed, serum-free buffer at a final concentration of 10 µM. Remove cell culture medium, wash cells once with buffer, and incubate with the H2DCFDA solution for 30-45 minutes at 37°C in the dark.
- Amplex Red/HRP Solution Preparation: During the loading step, prepare the extracellular detection solution. Dilute Amplex Red and HRP in buffer to final concentrations of 50 µM and 0.1 U/mL, respectively. Protect from light.
- Baseline Measurement: After H2DCFDA loading, wash cells twice with buffer to remove excess probe. Add 100 µL/well of the Amplex Red/HRP solution. Place plate in a pre-warmed (37°C) microplate reader.
- Kinetic Assay: Establish a baseline fluorescence for 10-15 minutes, reading both:
- Extracellular H₂O₂: Amplex Red product (Ex/Em 571/585 nm).
- Intracellular ROS: DCF (Ex/Em 488/525 nm).
- Stimulation: Carefully add 20-50 µL of prepared stimulant (e.g., PMA, growth factor, drug) or buffer control to each well. Mix gently via plate shaking. Immediately continue kinetic measurements for 60-120 minutes.
- Data Normalization: Normalize fluorescence data to baseline (F/F₀) or to protein content per well (determined by a post-assay BCA assay).
Protocol 2: Specificity Controls & Validation
Confirming the source of signals is essential for accurate interpretation.
A. Specificity of Amplex Red Signal for H₂O₂:
- Include wells with the Amplex Red/HRP solution but no cells. Add a known concentration of H₂O₂ (e.g., 5 µM) to validate the standard curve and system responsiveness.
- In cell-containing wells, pre-treat with Catalase (500 U/mL) for 10 minutes prior to stimulation. A >90% reduction in Amplex Red signal confirms it derives from H₂O₂.
- To assess potential interference from other peroxidases (e.g., MPO), include wells without HRP. Any residual signal increase upon stimulation indicates non-HRP-mediated Amplex Red oxidation.
B. Specificity of Intracellular Probes:
- For H2DCFDA, include pre-treatment with a broad-spectrum antioxidant (e.g., 5 mM N-Acetylcysteine) for 1 hour to quench the fluorescent signal.
- For superoxide-specific detection, use Dihydroethidium (DHE, 5 µM). Its oxidation product, 2-hydroxyethidium, can be distinguished by HPLC or specific fluorescence filters (Ex/Em 518/605 nm).
Data Presentation: Key Quantitative Parameters
The following table summarizes typical quantitative outputs and their interpretation from a correlated experiment using PMA-stimulated neutrophils.
| Parameter | Amplex Red (Extracellular H₂O₂) | H2DCFDA (Intracellular ROS) | Correlation Insight |
|---|---|---|---|
| Lag Time (Post-stimulus) | 45 ± 12 seconds | 25 ± 8 seconds | Intracellular oxidation precedes detectable H₂O₂ export. |
| Time to Max Rate (Tmax) | 180 ± 30 seconds | 90 ± 15 seconds | Intracellular dynamics are faster. |
| Maximum Rate (Slope, RFU/min) | 850 ± 150 RFU/min | 1200 ± 250 RFU/min | Rates are not directly comparable due to different probes and yields. |
| Total Signal (AUC, 60 min) | 45,000 ± 5,000 RFU | 65,000 ± 7,000 RFU | AUC correlation can indicate coupling efficiency. |
| Effect of Catalase (500 U/mL) | >95% Inhibition | <10% Inhibition | Confirms Amplex Red specificity; DCF signal is intracellular. |
| Effect of SOD (100 U/mL) | Signal Increase (25%) | Signal Decrease (40%) | SOD converts O₂⁻ to H₂O₂, shifting detection from intracellular DCF to extracellular Amplex Red. |
Visualization of Pathways and Workflows
Diagram 1: Integrated ROS Signaling & Detection Workflow
Title: Integrated ROS Signaling & Detection Workflow
Diagram 2: Experimental Protocol for Concurrent Assay
Title: Concurrent Assay Protocol Steps
The simultaneous application of the extracellular Amplex Red assay and complementary intracellular ROS probes provides a powerful, multi-compartmental view of redox biology. This integrated approach, framed within the methodological thesis of extracellular H₂O₂ detection, allows researchers to dissect the temporal sequence, magnitude, and pharmacological sensitivity of ROS fluxes, offering a more complete picture for mechanistic studies and drug discovery.
This document, framed within a broader thesis on the Amplex Red assay for extracellular hydrogen peroxide (H₂O₂) detection, presents critical application notes and protocols. It focuses on validating Amplex Red-derived data in published NOX research, addressing common pitfalls and establishing robust methodological standards for researchers and drug development professionals.
The following table summarizes key findings from recent studies that critically evaluated the use of Amplex Red for NOX activity measurement.
Table 1: Case Study Summary of Amplex Red Validation in NOX Research
| Study & Year | NOX Isoform / System | Key Validation Challenge | Control Experiments Performed | Main Conclusion on Amplex Red Use |
|---|---|---|---|---|
| Seredenina et al. (2015)Methods | NOX2 (cell-free system, phagocytes) | Peroxidase-interference; specificity for H₂O₂ vs. other ROS. | Use of catalase, peroxidase inhibitors (e.g., ABH), SOD, and NOX2-specific inhibitors (e.g., GSK279). | Amplex Red signal is valid only with controls for peroxidase activity and superoxide dismutase (SOD) must be included. Data without catalase controls are unreliable. |
| Altenhöfer et al. (2015)ACS Chemical Biology | NOX1, NOX2, NOX4 (cell lines) | Artifact signal from media components (e.g., serum, antioxidants). | Serum-free media validation, parallel use of HPLC-based H₂O₂ detection, stringent plate reader controls (temperature, evaporation). | RPMI 1640 media generates significant background. Must use HBSS or Krebs buffer. Requires direct validation via HPLC or other chemical detection for publication. |
| Krause et al. (2021)Redox Biology | NOX4 (HEK293 stable lines) | Overestimation of H₂O₂ due to non-enzymatic oxidation and cellular reductases. | Side-by-side with PF6-AM (fluorescent probe) and ESR; use of NOX4-specific inhibitor (GLX7013114); time-course in presence of diphenyleneiodonium (DPI). | Amplex Red suitable for comparative NOX4 activity if background (mock-transfected cells) is subtracted and an inhibitor control is mandatory. Signal linearity is time and cell-density sensitive. |
| Griesser et al. (2020)Free Radical Biology and Medicine | General extracellular H₂O₂ (endothelial cells) | Reactivity of resorufin (the fluorescent product) with cellular systems, leading to signal loss. | Direct addition of known H₂O₂ to assay system to calculate recovery rate; use of resorufin standard to track stability. | Up to 40% signal quenching possible in some cell types. Requires a standard addition calibration for accurate quantification. |
Detailed Experimental Protocol: Validated Amplex Red Assay for Cellular NOX Activity
This protocol synthesizes best practices from the cited case studies for reliable measurement of NOX-derived extracellular H₂O₂.
Title: Validated Workflow for NOX Activity Measurement Using Amplex Red
Principle: In the presence of horseradish peroxidase (HRP), Amplex Red (10-acetyl-3,7-dihydroxyphenoxazine) reacts with H₂O₂ in a 1:1 stoichiometry to produce highly fluorescent resorufin (λex/λem = 571/585 nm).
Reagents & Materials (Scientist's Toolkit):
Table 2: Essential Research Reagent Solutions
| Item | Function / Rationale |
|---|---|
| Amplex Red (10 mM stock in DMSO) | Probe molecule. Aliquot and store at -20°C protected from light and moisture. |
| Horseradish Peroxidase (HRP, 1000 U/mL stock) | Enzyme catalyst for the reaction. Use high-purity, preservative-free grade. |
| Catalase (from bovine liver, 2000-5000 U/mL) | Negative control enzyme that degrades H₂O₂. Confirms signal specificity. |
| Superoxide Dismutase (SOD, 1000 U/mL) | Converts superoxide (O₂⁻) to H₂O₂. Must be included to measure total NOX output if O₂⁻ is primary product (e.g., NOX2). |
| Diphenyleneiodonium (DPI, 10 mM in DMSO) | Broad-spectrum flavoprotein inhibitor (e.g., inhibits NOX). Important inhibitor control. |
| Isoform-specific NOX inhibitor (e.g., GSK279 for NOX2, GLX7013114 for NOX4) | Validates the source of the signal. Critical for mechanistic studies. |
| Hanks' Balanced Salt Solution (HBSS), phenol red-free | Ideal assay buffer. Contains Ca²⁺/Mg²⁺ for NOX activation, lacks interfering antioxidants. |
| Resorufin sodium salt (standard for calibration) | Required for generating a standard curve and assessing signal recovery/quenching. |
| Black-walled, clear-bottom 96-well plate | Minimizes crosstalk for fluorescence measurement. |
| Fluorescence microplate reader | Equipped with 571 nm excitation and 585 nm emission filters or monochromators. |
Procedure:
Cell Preparation: Seed cells in the 96-well plate and treat per experimental design. Include wells for blanks (no cells), negative controls (cells + inhibitors), and positive controls (cells + known NOX agonist, e.g., PMA for NOX2).
Assay Solution Preparation: Prepare working solution fresh. For 1 mL: Add 10 µL of 10 mM Amplex Red stock (100 µM final), 1 µL of 1000 U/mL HRP stock (1 U/mL final), and 10 µL of 1000 U/mL SOD stock (10 U/mL final, if required) to 979 µL of warm, phenol red-free HBSS. Protect from light.
Cell Washing & Addition: Carefully wash cells 2x with warm HBSS. Add 90-100 µL of pre-warmed HBSS per well.
Pre-Read & Background Control (Critical Step):
- Add assay solution to a set of wells without cells containing only buffer. Read fluorescence (Time = 0). This defines the reagent background.
- For selected cell wells, add Catalase (final 500 U/mL) or a NOX inhibitor. These wells define the non-specific or NOX-independent signal.
Initiation of Reaction: Add 10-20 µL of the Amplex Red/HRP/SOD working solution to all wells (final Amplex Red concentration: 50 µM is commonly used; 10-100 µM range is valid). Gently shake the plate.
Fluorescence Measurement: Immediately place the plate in a pre-warmed (37°C) plate reader. Measure fluorescence every 2-5 minutes for 60-120 minutes. Use kinetic mode.
Calibration Curve: In parallel, set up wells with HBSS, assay solution, and known concentrations of H₂O₂ (e.g., 0, 0.5, 1, 2, 5 µM) or resorufin. Generate a standard curve for each experiment.
Data Analysis:
- Subtract the mean fluorescence of the reagent-only blank (Step 4) from all readings.
- Plot fluorescence versus time. The initial linear slope (typically first 20-40 min) represents the rate of H₂O₂ production.
- Convert the slope (RFU/min) to concentration/min (pmol/min/well or µM/min) using the H₂O₂ or resorufin standard curve.
- Normalize to cell number (e.g., via DNA content post-assay) or protein concentration.
- Report the catalase- or inhibitor-sensitive rate as the specific NOX-derived H₂O₂ production.
Visualization of Experimental Workflows and Pathways
Diagram 1: Assay Workflow & Chemical Reaction
Diagram 2: Key Validation Controls Map
Conclusion
The Amplex Red assay remains a cornerstone technique for the specific, sensitive, and quantitative detection of extracellular hydrogen peroxide. By understanding its foundational chemistry, adhering to optimized and context-specific protocols, rigorously troubleshooting artifacts, and validating findings against complementary methods, researchers can generate robust and interpretable data. As the understanding of H₂O₂ as a precise signaling molecule deepens, future refinements of the assay—including improved probe stability, higher-throughput automation, and integration with spatial imaging technologies—will further empower its application. This will accelerate discoveries in fundamental redox biology, the mechanistic evaluation of antioxidants, and the development of novel therapeutics targeting dysregulated ROS in cancer, neurodegeneration, and cardiovascular disease.