From Disulfides to Sulfenic Acids: A Comprehensive Guide to Cysteine Oxidation State Analysis in Protein Research
Cysteine redox modifications are critical regulators of protein function, signaling, and disease.
From Disulfides to Sulfenic Acids: A Comprehensive Guide to Cysteine Oxidation State Analysis in Protein Research
Abstract
Cysteine redox modifications are critical regulators of protein function, signaling, and disease. This article provides a complete guide for researchers and drug development professionals on contemporary methods for assessing cysteine oxidation states. We cover the foundational biology of cysteine modifications, detail key experimental techniques from alkylation-based workflows to mass spectrometry and chemoproteomics, address common troubleshooting and optimization challenges, and offer a comparative analysis of method validation. This resource aims to empower scientists to select and implement the optimal strategy for their specific redox biology questions.
The Redox Landscape of Cysteine: Why Oxidation State Matters in Protein Function and Disease
Introduction to Cysteine as a Dynamic Redox Sensor in Proteins
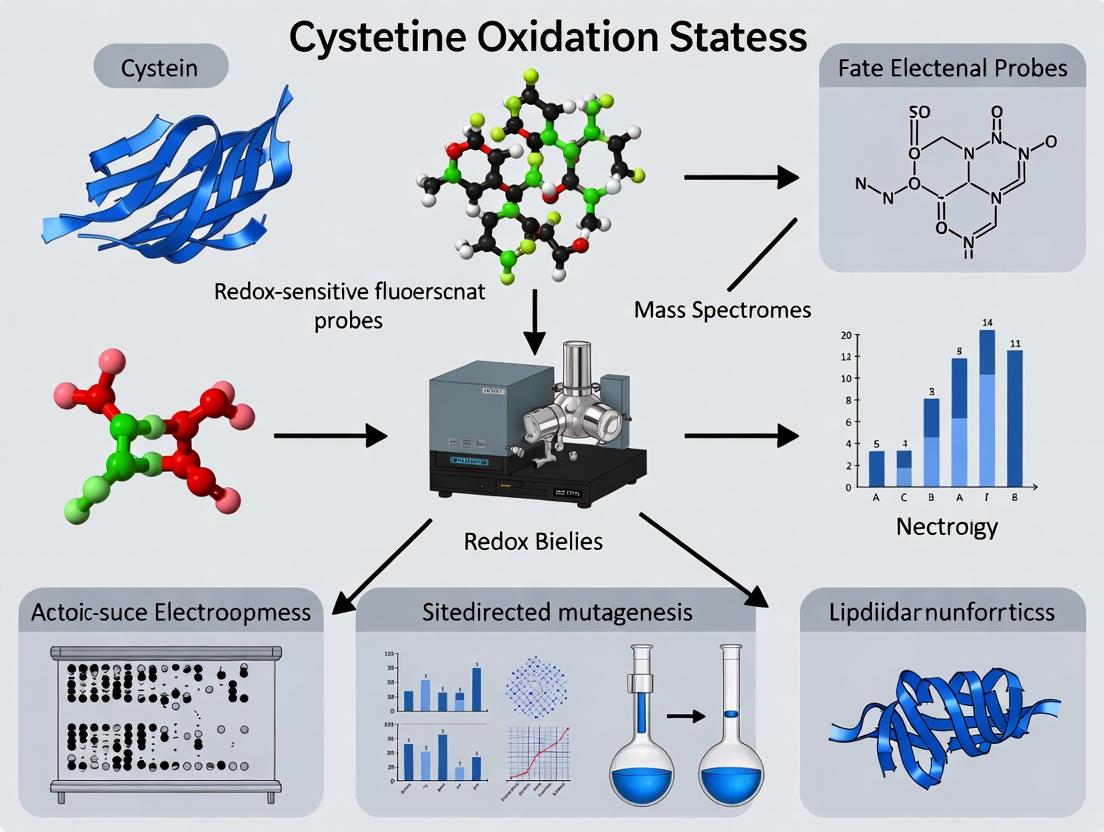
Cysteine residues in proteins are critical post-translational regulatory sites. Their thiol side chains undergo reversible oxidation to sulfenic acid (SOH), disulfide bonds (S-S), or glutathionylation (SSG) in response to cellular redox changes, acting as molecular switches that modulate protein function, localization, and stability. Accurate assessment of these states is foundational for understanding redox signaling and oxidative stress in disease and drug development.
Quantitative Data on Cysteine Redox Potentials and Reactivity
Table 1: Standard Redox Potentials and Modification Rates for Key Cysteine Oxidations
| Modification Type | Approximate E'° (mV vs. SHE)* | Typical Half-life | Key Detecting Probe |
|---|---|---|---|
| Free Thiol (SH) | -250 to -150 | Stable | Maleimides, Iodoacetamide |
| Sulfenic Acid (SOH) | N/A | Seconds to Minutes | Dimedone-based probes |
| Disulfide (S-S) | -150 to 0 | Stable until reduced | Reducing agents (DTT, TCEP) |
| S-Glutathionylation (SSG) | -150 to -70 | Minutes to Hours | Anti-glutathione antibodies |
| Sulfinic Acid (SO2H) | Irreversible | Stable | Specific antibodies |
| Sulfonic Acid (SO3H) | Irreversible | Stable | Specific antibodies |
*SHE = Standard Hydrogen Electrode; Potentials are environment-dependent.
Table 2: Common Biophysical Methods for Cysteine Oxidation State Analysis
| Method | Detection Principle | Sensitivity | Throughput | Key Limitation |
|---|---|---|---|---|
| Mass Spectrometry (MS) | Mass shift from modifications | High (fmol) | Low-Medium | Artifacts during prep |
| Biotin Switch Assay | Selective labeling of SNO | Moderate | Low | Requires specific controls |
| OxICAT | Isotopic thiol trapping | High | Low | Technically complex |
| CPM / IAM-based assays | Fluorescence from free thiols | Moderate | High | Detects only reduced thiols |
| Redox Western Blot | Electrophoretic mobility shift | Low-Moderate | Medium | Limited to specific proteins |
Experimental Protocols
Protocol 1: The Biotin-Switch Technique for S-Nitrosylation (BSN) Detection
Adapted from Jaffrey & Snyder (2001) for general redox cysteine profiling.
Objective: To selectively identify S-nitrosylated (SNO) or, with modifications, sulfenylated cysteine residues.
Reagents & Solutions:
- HENS Buffer: 250 mM HEPES-NaOH pH 7.7, 1 mM EDTA, 0.1 mM Neocuproine, 1% SDS.
- Blocking Buffer: HENS with 2.5% SDS and 20 mM Methyl Methanethiosulfonate (MMTS).
- Reducing Buffer: HENS with 1% SDS and 10 mM Ascorbate (for SNO) or 1 mM Arsenite (for SOH).
- Labeling Buffer: HENS with 1% SDS, 4 mM Biotin-HPDP (or PEG-maleimide for free thiols).
Procedure:
- Cell Lysis: Lyse cells/tissue in HENS buffer + protease inhibitors. Clarify by centrifugation.
- Free Thiol Blocking: Incubate lysate with Blocking Buffer (MMTS) at 50°C for 20 min with agitation. This alkylates all free, reduced thiols.
- Precipitation: Remove excess MMTS by acetone precipitation. Resuspend pellet in HENS.
- Selective Reduction: Treat samples with Reducing Buffer. For SNO detection: Use Ascorbate (10 mM, 1 hr, RT) to selectively reduce S-NO bonds. For SOH detection: Use Sodium Arsenite (1 mM).
- Biotinylation: Add Biotin-HPDP to the Labeling Buffer and incubate with sample for 1 hr at RT. This labels the newly reduced thiols.
- Detection: Precipitate protein, resuspend, and perform streptavidin pulldown. Analyze by Western blot or MS.
Protocol 2: OxICAT (Oxidative Isotope-Coded Affinity Tag)
Adapted from Leichert et al., 2008.
Objective: To quantitatively measure the in vivo redox state of cysteine thiols on a proteome-wide scale.
Procedure:
- In Vivo Quenching & Lysis: Rapidly lyse cells in an acidic, denaturing lysis buffer (e.g., 100 mM Tris, 1% SDS, pH 4.5, 40 mM chloroacetamide) to "freeze" the redox state.
- Differential Labeling:
- Divide lysate into two aliquots.
- Light Labeling (Reduced Thiols): Treat first aliquot with a "light" (12C) version of Iodoacetamide (ICAT reagent) to alkylate all reduced thiols present at quenching.
- Reduction & Heavy Labeling (Oxidized Thiols): Fully reduce the second aliquot with TCEP (10 mM, 1 hr). Then alkylate with the "heavy" (13C) ICAT reagent. This labels thiols that were initially oxidized.
- Mixing & Digestion: Combine the light- and heavy-labeled samples in a 1:1 ratio. Digest with trypsin.
- Affinity Purification: Isolate ICAT-labeled peptides using the biotin tag on the ICAT reagent (streptavidin beads).
- LC-MS/MS Analysis: Analyze by liquid chromatography-tandem mass spectrometry. The relative peak intensities of the light (reduced) and heavy (oxidized) peptide pairs provide a direct quantitative ratio of the redox state of each cysteine at the time of quenching.
Visualizations
Diagram 1: Cysteine Redox Modification Cycle
Diagram 2: Biotin-Switch Assay Workflow
Diagram 3: OxICAT Quantitative Proteomics Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
| Reagent / Kit | Primary Function in Redox Assessment |
|---|---|
| Iodoacetamide (IAM) & Derivatives | Alkylates free thiols to prevent further oxidation during sample prep. Iodoacetyl-PEG-biotin enables detection. |
| Methyl Methanethiosulfonate (MMTS) | Membrane-permeable thiol blocker used in Biotin-Switch assays to rapidly cap free cysteines. |
| Biotin-HPDP | Thiol-reactive, cleavable biotinylation reagent. HPDP group reacts with SH, biotin enables affinity capture. |
| TCEP (Tris(2-carboxyethyl)phosphine) | Strong, odorless reducing agent to reduce disulfides. More stable than DTT in acidic conditions. |
| Dimedone & DCP-Bio1 | Specific, nucleophilic probes that covalently tag sulfenic acid (SOH) formations. |
| Anti-Glutathione Antibody | For direct immunodetection of protein S-glutathionylation (SSG) via Western blot or immunofluorescence. |
| ICAT Reagents (Isotope-Coded Affinity Tags) | Paired light/heavy isotopes for MS-based quantification of redox states, as used in OxICAT. |
| CPM (7-Diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin) | Thiol-sensitive fluorescent dye; fluorescence increases upon reaction with free SH groups. |
| Sodium Ascorbate | Selective reducing agent for S-nitrosothiols (SNO) with minimal effect on disulfides. |
| PMSF & Protease Inhibitor Cocktails | Essential to prevent artefactual cysteine modifications by proteolytic degradation during lysis. |
Understanding the diverse oxidation states of cysteine residues is central to the thesis "Methods for assessing cysteine oxidation states in proteins research." This application note details the biological significance, detection methods, and experimental protocols for analyzing key cysteine modifications—from reversible redox signaling events like sulfenic acid formation and disulfide bonding to irreversible over-oxidation to sulfinic and sulfonic acids. These states are critical in enzyme catalysis, structural stabilization, and cellular redox signaling, with direct implications for drug development targeting oxidative stress-related diseases.
Table 1: Key Cysteine Oxidation States and Their Properties
| Oxidation State | Chemical Formula | Reversibility | Typical pKa (of precursor) | Key Detection Methods | Biological Role |
|---|---|---|---|---|---|
| Thiol (Reduced) | -SH | N/A | ~8.5 | Ellman's assay, Maleimide probes | Enzyme active site, Metal binding |
| Disulfide | -S-S- | Reversible (via reductants) | N/A | Non-reducing SDS-PAGE, Mass Spectrometry | Structural stability, Redox buffering |
| Sulfenic Acid | -SOH | Reversible (via thiols) | ~6.5 | Dimedone-based probes, MS | Redox signaling, Enzyme regulation |
| Sulfinic Acid | -SO2H | Irreversible (in mammals) | ~2.5 | Specific antibodies, MS | Oxidative stress marker |
| Sulfonic Acid | -SO3H | Irreversible | <1 | MS, Amino acid analysis | Terminal oxidation, Damage marker |
Application Notes & Protocols
Protocol 1: Trapping and Detection of Protein Sulfenic Acids
Principle: Sulfenic acids are transient and reactive. Nucleophilic probes like 5,5-dimethyl-1,3-cyclohexanedione (dimedone) selectively react with sulfenic acid, forming a stable thioether adduct for downstream analysis.
Detailed Methodology:
- Cell Lysis: Lyse cells or tissue in degassed lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40) supplemented with 10 mM dimedone probe (e.g., DCP-Bio1 or DYn-2) and protease inhibitors. Include 10 mM N-ethylmaleimide (NEM) to block free thiols after dimedone trapping if necessary.
- Trapping Reaction: Incubate lysate at 25°C for 1 hour with gentle rotation.
- Protein Clean-up: Precipitate proteins using cold acetone, wash twice, and resuspend in appropriate buffer.
- Detection:
- Biotinylated Probes: Pull-down with streptavidin beads, wash, elute with SDS-PAGE sample buffer, and analyze by Western blot.
- Fluorescent Probes: Visualize directly by in-gel fluorescence scanning.
- Mass Spectrometry: Digest proteins with trypsin, enrich biotinylated peptides with streptavidin, and analyze by LC-MS/MS to identify modification sites.
Protocol 2: Differentiating Reversible Disulfides from Irreversible Oxidation
Principle: Sequential alkylation with thiol-reactive reagents of different masses allows differentiation of reduced, reversibly oxidized, and irreversibly oxidized cysteines via mass spectrometry.
Detailed Methodology (SP3-Rox Method):
- Sample Preparation: Denature protein extract in 1% SDS, 100 mM Tris-HCl, pH 7.5.
- Block Free Thiols (Step 1): Add 20 mM iodoacetamide (IAM, light, +57 Da) to alkylate all reduced cysteine thiols. Incubate 30 min in the dark at 25°C.
- Reduce Reversible Oxidations: Add 10 mM Tris(2-carboxyethyl)phosphine (TCEP) to reduce disulfides and sulfenic acids. Incubate 30 min at 37°C.
- Label Newly Reduced Thiols (Step 2): Add 20 mM isotopically heavy N-ethylmaleimide (d5-NEM, heavy, +5 Da) to alkylate cysteines that were reversibly oxidized. Incubate 30 min in the dark at 25°C.
- Clean-up & Analysis: Quench reaction, clean proteins via SP3 beads, digest with trypsin, and analyze by LC-MS/MS. Peptide spectra reveal oxidation state: IAM-only (irreversible SO2/SO3), IAM+d5-NEM (reversible oxidation), d5-NEM only (reduced thiol).
Protocol 3: Immunoblotting for Sulfinic and Sulfonic Acids
Principle: While mass spectrometry is comprehensive, immunoblotting offers rapid validation. Antibodies against sulfonic acid (e.g., anti-cysteic acid) and specific sulfinic acid (e.g., anti-Prx-SO2/3) are available.
Detailed Methodology:
- Sample Preparation: Resuspend proteins in non-reducing Laemmli buffer (without β-mercaptoethanol or DTT) to preserve oxidation states.
- Electrophoresis: Run samples on standard SDS-PAGE.
- Transfer & Blocking: Transfer to PVDF membrane, block with 5% BSA in TBST for 1 hour.
- Primary Antibody Incubation: Incubate with anti-sulfinic/sulfonic acid primary antibody (e.g., 1:1000 dilution) overnight at 4°C.
- Detection: Follow with HRP-conjugated secondary antibody and chemiluminescent detection. Note: Always include positive (e.g., over-oxidized peroxiredoxin) and negative (reduced) controls.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Cysteine Redox Research
| Reagent | Function & Specific Example |
|---|---|
| Dimedone-based Probes (e.g., DCP-Bio1) | Chemoselectively traps sulfenic acids for enrichment or visualization. |
| Isotopically Coded Alkylating Agents (Light/Heavy IAM, NEM) | Labels cysteine thiols of different oxidation states for quantitative MS. |
| Strong Reductants (TCEP, DTT) | Reduces reversible disulfides; TCEP is preferred for pH stability and lack of thiol generation. |
| Thiol Blockers (Iodoacetamide, NEM) | Alkylates free thiols to prevent post-lysis artifacts. |
| Sulfinic Acid Antibodies (e.g., anti-Prx-SO2/3) | Enables specific detection of over-oxidized peroxiredoxins by Western blot. |
| Biotin-HPDP | Thiol-disulfide exchange reagent for labeling reduced thiols or measuring redox state. |
| Streptavidin Beads | Enriches biotin-tagged proteins/peptides from complex mixtures. |
| Redox-sensitive GFP (roGFP) | Genetically encoded biosensor for real-time imaging of cellular glutathione redox potential. |
Visualization of Pathways and Workflows
Title: Reversible and Irreversible Cysteine Oxidation Pathways
Title: SP3-ROX MS Workflow for Cysteine States
Application Notes
This document provides methodological support for the investigation of cysteine oxidation states, a critical determinant in redox signaling, allosteric control, and structural integrity of proteins. Accurate assessment of these states is foundational for research in cellular signaling, enzymology, and therapeutic development targeting oxidative stress pathways.
1. Redox Signaling: Reversible cysteine oxidation (e.g., to sulfenic acid, S-nitrosothiols, or glutathionylated disulfides) forms the basis of hydrogen peroxide and nitric oxide signaling networks. Quantifying these labile modifications in response to stimuli is essential for mapping signaling dynamics.
2. Allosteric Regulation: The formation or reduction of disulfides can induce long-range conformational changes, modulating protein function. Methods to trap and characterize these states are required to elucidate allosteric mechanisms.
3. Structural Disulfide Bonds: Irreversible, stable disulfides are crucial for extracellular protein folding and stability. Differentiating these from regulatory disulfides is a key analytical challenge.
Table 1: Common Cysteine Oxidative Modifications and Detection Strategies
| Modification (R-S-) | Typical Role | Key Detection Method(s) | Chemical Lability |
|---|---|---|---|
| Sulfhydryl (Thiol) | Reduced, active site | Maleimide labeling, DTNB assay | Stable if reduced |
| Sulfenic Acid | Redox sensing | Dimedone-based probes (e.g., DYn-2) | Highly reactive |
| Intra/Inter Disulfide | Structural / Regulatory | Non-reducing PAGE, Diagonal Electrophoresis | Reducible (DTT) |
| S-Glutathionylation | Redox regulation | Biotinylated glutathione ethyl ester (BioGEE), Anti-GSH antibodies | Reducible, labile |
| S-Nitrosylation | NO signaling | Biotin switch technique (BST), SNO-RAC | Photolabile, Cu²⁺-sensitive |
| Sulfinic/Sulfonic Acid | Irreversible oxidation | Antibodies (e.g., anti-Cys-SO₂H/SO₃H) | Irreversible |
Table 2: Quantitative Comparison of Mass Spectrometry (MS) Approaches for Cysteine Oxidation
| MS Method | Principle | Resolution | Throughput | Key Requirement |
|---|---|---|---|---|
| ICAT (Isotope-Coded Affinity Tag) | Light/heavy tags on free thiols | Quantitative, comparative | Medium | Thiol alkylation at specific time point |
| OxMRM (Oxidation-specific Multiple Reaction Monitoring) | Targeted MS/MS of specific peptides | High sensitivity & precision | High | Pre-defined peptide transitions |
| IPTL (Isobaric Protein Terminal Labeling) | Tandem mass tags post-digestion | Multiplexing (up to 16-plex) | High | Efficient protein labeling |
| Redox-DIGE (2D-Difference Gel Electrophoresis) | Fluorescent CyDyes on thiols pre-separation | Comparative, visual | Low-Medium | Specialized dye chemistry |
| CPT (Cysteine Reactivity Profiling) | Probes with alkyne/azide handles for click chemistry | Activity-based profiling | High | Functionalized probe design |
Detailed Protocols
Protocol 1: Trapping and Enrichment of Sulfenic Acid Modifications Using Dyn-2 Objective: To selectively label and enrich protein sulfenic acid modifications from cell lysates. Materials: DYn-2 (alkyne-functionalized 1,3-cyclohexanedione derivative), Dimethyl sulfoxide (DMSO), Copper(II) sulfate, Tris(2-carboxyethyl)phosphine (TCEP), TBTA ligand, Azide-functionalized biotin, Streptavidin beads, Lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, with fresh 20 mM N-ethylmaleimide (NEM) and protease inhibitors). Procedure:
- Treat cells with experimental stimulus (e.g., H₂O₂) or vehicle control.
- Lyse cells in ice-cold lysis buffer containing NEM to alkylate free thiols. Centrifuge (16,000 x g, 15 min, 4°C).
- Incubate clarified lysate with 100 µM DYn-2 (from 10 mM stock in DMSO) for 1 hour at room temperature, protected from light.
- Perform click chemistry: To the lysate, add 100 µM biotin-azide, 1 mM TCEP, 100 µM TBTA, and 1 mM CuSO₄. Incubate with rotation for 1 hour at room temperature.
- Precipitate proteins using methanol/chloroform to remove excess reagents. Resuspend pellet in PBS with 1% SDS.
- Enrich: Dilute samples to 0.1% SDS, incubate with pre-washed streptavidin beads for 2 hours at 4°C.
- Wash beads stringently (e.g., with 1% SDS, 4 M urea, and high-salt buffers).
- Elute by boiling in Laemmli buffer with 20 mM DTT for analysis by immunoblot or MS.
Protocol 2: Diagonal Electrophoresis for Mapping Regulatory Disulfide Bonds Objective: To separate peptides containing disulfide-linked complexes from non-linked peptides. Materials: Non-reducing Laemmli buffer (no DTT/β-mercaptoethanol), Horizontal electrophoresis system, Glass plates for second dimension, Performic acid oxidation solution (1:9 30% H₂O₂: 88% formic acid, incubated 1 hour at RT before use), Standard SDS-PAGE reagents. Procedure:
- Separate protein(s) of interest by non-reducing SDS-PAGE in the first dimension.
- Excise the lane, incubate in 50 mL of performic acid oxidation solution for 2 hours on ice to oxidize and cleave all disulfides to cysteic acid.
- Neutralize the gel strip by washing in several changes of electrophoresis buffer.
- Embed the strip horizontally on top of a second-dimension SDS-PAGE gel (now containing a reducing agent if desired).
- Run the second dimension. Peptides not originally connected by a disulfide will run on the diagonal. Peptides that were disulfide-linked will now be smaller/more acidic and migrate below the diagonal.
- Visualize by Coomassie or silver stain. Spots below the diagonal represent former disulfide-linked partners.
Protocol 3: The Biotin Switch Technique (BST) for S-Nitrosylation Objective: To selectively detect S-nitrosylated proteins. Materials: Methyl methanethiosulfonate (MMTS), NeutrAvidin or Streptavidin beads, Biotin-HPDP (N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide), HEN buffer (250 mM HEPES pH 7.7, 1 mM EDTA, 0.1 mM Neocuproine). Procedure:
- Lyse samples in HEN buffer with 1% CHAPS. Block free thiols by adding MMTS to 20 mM and incubating at 50°C for 20 min with frequent vortexing.
- Precipitate proteins with acetone to remove MMTS. Resuspend in HEN buffer with 1% SDS.
- Reduce S-NO bonds to thiols by adding ascorbate (sodium salt) to a final concentration of 20 mM and incubating for 1 hour at room temperature.
- Label the newly revealed thiols with 0.4 mM Biotin-HPDP (from stock in DMSO) for 1 hour at room temperature.
- Precipitate proteins again to remove excess biotin. Resuspend and incubate with NeutrAvidin beads overnight at 4°C.
- Wash beads thoroughly and elute bound proteins with Laemmli buffer containing DTT for downstream analysis.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Kit | Primary Function | Key Consideration |
|---|---|---|
| Iodoacetyl Tandem Mass Tag (iodoTMT) | Multiplexed (6-plex) isotopic labeling of reduced cysteine thiols for quantitative MS. | Alkylates free thiols; blocking step is critical. |
| PEG Maleimide (PEG-Mal) | Molecular weight shift assay for free thiols via non-reducing gel. | Different PEG sizes (e.g., 5 kDa) can be used. |
| Anti-Sulfenic Acid Antibodies | Immunoblot detection of some stabilized sulfenic acid modifications. | Specificity varies; requires careful validation. |
| Mono-bromobimane (mBBr) | Thiol-specific fluorescent label for gel-based or HPLC detection. | Useful for quantifying thiol/disulfide ratios. |
| CPM (7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin) | Fluorescent dye for monitoring thiol availability/reactivity in real-time. | Used in kinetic assays of protein unfolding/folding. |
| Redox-Sensitive GFP (roGFP) | Genetically encoded biosensor for real-time imaging of glutathione redox potential in live cells. | Must be targeted to specific cellular compartments. |
| Cy5/Cy3 Maleimide Dyes | For Redox 2D-DIGE, differential labeling of control vs. treated thiol pools. | Requires specialized imaging systems. |
Visualizations
Diagram 1: Cysteine Redox Signaling Pathway Workflow
Diagram 2: Experimental Workflow for Redox Proteomics
Diagram 3: Logic of Cysteine Oxidation State Assessment
Application Notes & Protocols
Within the broader thesis investigating methods for assessing cysteine oxidation states in proteins, this document provides specific application notes and protocols. Understanding these oxidative modifications is critical, as they are a primary mechanistic link between oxidative stress and major disease pathologies, including neurodegeneration, cancer, and aging.
Table 1: Key Cysteine Oxidative Modifications and Their Disease Associations
| Modification | Chemical Formula/Description | Associated Disease Context | Quantitative Change (Example) |
|---|---|---|---|
| Sulfenic Acid | R-SOH | Reversible signaling in cancer cell proliferation; transient in neurodegeneration. | Up to ~40% increase in specific protein sulfenylation under H₂O₂ stress (e.g., PTP1B). |
| Disulfide Bond | R-S-S-R' | Protein misfolding in AD/PD; redox regulation in tumor suppressors (e.g., PTEN). | Can exceed 60% of total protein pool in ER stress models. |
| S-Glutathionylation | R-S-SG | Protective in acute stress; dysregulated in CVD, aging. | Levels can rise from basal ~5% to >30% post-oxidative insult. |
| Sulfinic Acid | R-SO₂H | Largely irreversible; marker of severe stress in neurodegeneration. | Quantification is low (<2% typically) but persistent. |
| S-Nitrosylation | R-SNO | Neuroprotective or toxic in ND; pro-/anti-tumorigenic in cancer. | nM to µM concentrations in tissue samples; highly variable. |
Protocol 1: Biotin-Switch Technique for S-Nitrosylation Mapping Objective: To selectively label and detect S-nitrosylated proteins from complex lysates.
- Cell Lysis & Blocking: Lyse tissue/cells in HEN buffer (250 mM HEPES pH 7.7, 1 mM EDTA, 0.1 mM Neocuproine) with 2.5% SDS. Add Methyl methanethiosulfonate (MMTS) to 20 mM to block free thiols. Incubate at 50°C for 20 min with agitation.
- Acetone Precipitation: Remove excess MMTS by adding 2 volumes of pre-chilled acetone, incubate at -20°C for 20 min, pellet protein, and wash twice with 70% acetone.
- Nitrosothiol Reduction & Biotinylation: Resuspend pellet in HENS buffer (HEN + 1% SDS). For every 1 mg of protein, add 1 mM ascorbate and 4 mM biotin-HPDP. Incubate at 25°C for 1 hour in the dark.
- Pull-down & Analysis: Remove unreacted biotin-HPDP via acetone precipitation. Resuspend in neutralization buffer. Incubate with streptavidin-agarose beads for 1 hour. Wash beads, elute with SDS-PAGE loading buffer containing β-mercaptoethanol, and analyze by immunoblotting.
Protocol 2: Dimedone-Based Probe for Sulfenic Acid Detection Objective: To detect protein sulfenylation in situ.
- Probe Incubation: Treat live cells or freshly lysed tissue with a cell-permeable, tagged dimedone derivative (e.g., DYn-2 or biotin-conjugated dimedone) at 10-100 µM in serum-free media/PBS. Incubate for 30-60 min at 37°C.
- Cell Lysis & Capture: Wash cells and lyse in non-reducing, non-denaturing lysis buffer. For biotinylated probes, incubate lysate with streptavidin beads overnight at 4°C.
- Washing & Elution: Wash beads stringently (e.g., high salt, low detergent). Elute bound proteins with Laemmli buffer containing 10 mM DTT.
- Downstream Analysis: Analyze eluates by SDS-PAGE and immunoblotting for targets of interest, or by mass spectrometry for global profiling.
The Scientist's Toolkit: Key Reagents for Cysteine Redox Profiling
| Reagent | Function & Application |
|---|---|
| Iodoacetyl Tandem Mass Tag (iodoTMT) | Isobaric tags for multiplexed quantification of reduced cysteine thiols. |
| N-ethylmaleimide (NEM) | Thiol-alkylating agent to block free cysteines and "lock" the redox state during lysis. |
| Biotin-HPDP | Thiol-reactive, biotinylated probe used in the biotin-switch technique for SNO detection. |
| Dimedone-based probes (e.g., DYn-2) | Cyclic 1,3-diketones that selectively and covalently react with sulfenic acids. |
| Streptavidin Agarose/Magnetic Beads | For affinity purification of biotinylated proteins post-probe labeling. |
| Ascorbate (Vitamin C) | Specific reducing agent for S-nitrosothiols in the biotin-switch protocol. |
| DTT (Dithiothreitol) / TCEP | General reducing agents to reduce disulfides; used as controls or for elution. |
| Antibody against Glutathione | For immunodetection of protein S-glutathionylation. |
Diagram 1: Cysteine Oxidation in Disease Pathways
Diagram 2: Biotin-Switch Technique Workflow
The assessment of cysteine oxidation states in proteins is a cornerstone of redox biology research, with direct implications for understanding disease mechanisms and developing targeted therapeutics. This field rests on three fundamental pillars: the Redox Potential (Eₕ) of the cellular environment and individual proteins, the physiologically distinct Cellular Compartments that maintain unique redox milieus, and the Reversibility of oxidative modifications that underpins redox signaling. This article provides detailed application notes and experimental protocols for researchers investigating these concepts within drug development and mechanistic studies.
Core Conceptual Frameworks & Quantitative Data
Standard Redox Potentials of Key Cysteine Modifications
The thermodynamic tendency of a thiol to become oxidized is governed by its redox potential. The following table summarizes standard potentials for biologically relevant couples.
Table 1: Standard Reduction Potentials for Cysteine Redox Couples
| Redox Couple (Reduced Oxidized) | Approximate E'⁰ (mV) at pH 7.0, 25°C | Biological Relevance |
|---|---|---|
| 2GSH GSSG + 2H⁺ + 2e⁻ | -240 | Glutathione redox buffer; cellular background potential |
| Protein Cys-SH Protein Cys-SOH + 2H⁺ + 2e⁻ | -150 to +150 | Sulfenic acid formation; highly dependent on protein microenvironment |
| Protein Cys-SH Protein Cys-S-S-Cys-Protein + 2H⁺ + 2e⁻ | -180 to -330 | Intramolecular/protein disulfide formation |
| Protein Cys-SH Protein Cys-S-SG + 2H⁺ + 2e⁻ | -150 to -200 | S-glutathionylation; mixed disulfide |
| Trx-(SH)₂ Trx-S₂ + 2H⁺ + 2e⁻ | -270 to -290 | Thioredoxin system; key reductant |
Compartmentalized Redox Environments in Mammalian Cells
The redox state is not uniform within the cell. Key compartments maintain distinct glutathione redox potentials (Eₕc), which dictate the stability of cysteine modifications.
Table 2: Compartment-Specific Glutathione Redox Potentials (Eₕc)
| Cellular Compartment | [GSH]:[GSSG] Ratio (Approx.) | Glutathione Redox Potential (Eₕc) (mV) | pH | Implications for Cysteine Oxidation |
|---|---|---|---|---|
| Cytosol / Nucleus | 100:1 to 300:1 | -260 to -280 | ~7.2 | Reducing; disulfides generally unstable, signaling modifications are transient. |
| Mitochondrial Matrix | 20:1 to 40:1 | -280 to -300 | ~8.0 | Highly reducing despite lower ratio due to alkaline pH; maintains metabolic enzyme thiols. |
| Endoplasmic Reticulum Lumen | 1:1 to 3:1 | -180 to -210 | ~7.1 | Oxidizing; favors formation and isomerization of structural disulfide bonds in secretory proteins. |
| Extracellular Space / Secreted | 1:30 to 1:100 | -120 to -150 | ~7.4 | Highly oxidizing; favors stabilized disulfides, modifications are often irreversible. |
| Lysosome Lumen | Not well defined | Estimated more oxidizing | ~4.5-5.0 | Acidic pH affects thiol pKa and stability of modifications. |
Experimental Protocols
Protocol: Assessing Compartment-Specific Redox Potential Using roGFP2 Probes
Objective: To measure the glutathione-dependent redox potential (Eₕc) within specific organelles (e.g., cytosol, mitochondria) in live cells. Principle: Redox-sensitive Green Fluorescent Protein 2 (roGFP2) is a genetically encoded sensor. Oxidation causes a reversible excitation peak shift, measurable by ratiometric fluorescence.
Materials: See Scientist's Toolkit, Section 5.
Procedure:
- Sensor Expression: Transfect cells with a plasmid encoding roGFP2 targeted to your compartment of interest (e.g., roGFP2-Mito for mitochondria, roGFP2-ER).
- Cell Preparation: 24-48h post-transfection, seed cells into a black-walled, clear-bottom 96-well plate or prepare on imaging dishes. Allow to adhere overnight.
- Ratiometric Measurement:
- For a plate reader: Acquire fluorescence intensities using two excitation wavelengths (Ex 400nm ± 10nm and Ex 490nm ± 10nm) with a single emission (Em 510nm ± 10nm).
- For microscopy: Capture two images per cell using 405nm and 488nm excitation and a 510/20nm emission filter.
- Calibration & Quantification (In situ):
- After baseline read, treat cells with 10mM DTT (fully reduced control) for 15 min and measure.
- Wash cells and treat with 1-5mM Diamide (oxidizing agent) for 15 min and measure.
- Calculate the ratiometric value (R) = Intensity(Ex400)/Intensity(Ex490).
- Calculate the degree of oxidation: Oxidation = (R - R₍DTT₎) / (R₍Diamide₎ - R₍DTT₎).
- Convert to Eₕc using the Nernst equation: Eₕc = E⁰ - (RT/nF)ln([GSH]²/[GSSG]), where the sensor's E⁰ is -280 mV for roGFP2-Orp1. Use established calibration curves linking ratio to Eₕc.
- Experimental Treatment: Treat cells with your experimental stimulus (e.g., H₂O₂, drug, nutrient stress) and perform ratiometric measurements over time.
Protocol: Trapping and Identifying Reversible Cysteine Oxidations (Biotin Switch Technique, BST)
Objective: To selectively label and isolate proteins containing reversibly oxidized (S-nitrosylated or S-sulfenylated) cysteines. Principle: Free thiols are blocked, labile oxidative modifications are selectively reduced, and the newly revealed thiols are tagged with a biotinylated agent for affinity purification.
Procedure:
- Cell Lysis and Blocking: Lyse cells in HEN buffer (25mM HEPES, 1mM EDTA, 0.1mM Neocuproine) + 2.5% SDS. Immediately add the alkylating agent Methyl Methanethiosulfonate (MMTS, 20-50mM final) to block all free thiols. Incubate at 50°C for 20 min with frequent vortexing.
- Precipitation: Remove excess MMTS by acetone precipitation (2-3 volumes). Resuspend pellet in HEN buffer with 1% SDS.
- Selective Reduction: For S-nitrosothiols: Add ascorbate (1mM final). For S-sulfenic acids: Use arsenite (1-5mM) or dimedone-based probes. Incubate at room temperature for 1 hour. Note: Negative controls omit the reducing agent.
- Biotin Tagging: Add a thiol-reactive biotinylation reagent (e.g., EZ-Link HPDP-Biotin or Biotin-HPDP at 0.5-1mM, or iodoacetyl-PEG₂-biotin at 0.25mM). Incubate at room temperature for 1-3h in the dark.
- Affinity Capture: Remove excess biotin by acetone precipitation. Resuspend pellet in neutralization buffer. Incubate with pre-equilibrated streptavidin-agarose beads overnight at 4°C.
- Washing and Elution: Wash beads stringently (e.g., with high-salt, low-detergent buffers). Elute bound proteins directly with Laemmli buffer containing 100mM DTT (reduces biotin-thiol bond) for Western blot analysis or perform on-bead trypsin digestion for mass spectrometry identification.
Diagrams: Pathways and Workflows
Cysteine Oxidation States & Reversibility Pathway
Diagram 1: Reversible and Irreversible Cysteine Oxidation Fates
Experimental BST Workflow for Reversible Oxidation
Diagram 2: Biotin Switch Technique (BST) Workflow
The Scientist's Toolkit
Table 3: Key Research Reagent Solutions for Cysteine Redox Studies
| Reagent / Material | Function & Role in Experiment | Example / Notes |
|---|---|---|
| roGFP2 Plasmids | Genetically encoded sensor for ratiometric measurement of Eₕc. | roGFP2 (cytosol), roGFP2-Orp1 (H₂O₂ specific), mito-roGFP2, ER-roGFP2. Available from Addgene. |
| DTT (Dithiothreitol) | Strong reducing agent. Used to fully reduce samples (calibration/control). | 10-100mM in aqueous solution. Unstable at high pH. |
| Diamide (Azodicarboxylate) | Thiol-oxidizing agent. Used to fully oxidize samples (calibration/control). | Typically 1-5mM in cells. Induces disulfide formation. |
| MMTS (Methyl Methanethiosulfonate) | Membrane-permeable, thiol-specific alkylating agent. Blocks free -SH groups in BST. | 20-50mM in lysis buffer. Prevents artifactual oxidation during processing. |
| HPDP-Biotin (N-(6-(Biotinamido)hexyl)-3'-(2'-pyridyldithio)propionamide) | Thiol-reactive, cleavable biotinylation reagent. Tags newly reduced cysteines in BST. | Forms disulfide bond with thiol, allowing elution with DTT. |
| Streptavidin Agarose Beads | High-affinity capture resin for biotinylated proteins. Used to isolate tagged proteins in BST. | High binding capacity (>1 µg biotinylated protein per µL beads). |
| Iodoacetyl-PEG₂-Biotin | Thiol-alkylating biotin reagent. Irreversibly labels thiols via iodoacetamide chemistry. | Alternative to HPDP-Biotin for irreversible capture (not BST-specific). |
| Dimedone & Derivatives (e.g., DCP-Bio1) | Chemoselective probe that covalently tags sulfenic acids (-SOH). Used to detect this transient state. | Can be conjugated to biotin or fluorescent tags for detection/enrichment. |
| Anti-Glutathione Antibody | Detects protein-bound glutathione (S-glutathionylation) in Western blot or immunofluorescence. | Allows specific detection of this mixed disulfide modification. |
Toolkit for Detection: Step-by-Step Methods for Profiling Cysteine Oxidation
Within the broader thesis on "Methods for assessing cysteine oxidation states in proteins," a foundational and critical step is the rapid and irreversible chemical trapping of labile cysteine modifications. Cysteine residues can exist in various oxidation states (e.g., sulfenic acid (-SOH), persulfide (-SSH), in reversible disulfides) that are highly reactive and transient. To accurately capture this redox proteome snapshot, alkylating agents like iodoacetamide (IAA), N-ethylmaleimide (NEM), and iodoacetic acid (IAA) are employed. These reagents covalently bind to free thiols (-SH), blocking further oxidation or reduction during sample processing, thereby "locking in" the redox state at the moment of lysis.
Application Notes
1. Reagent Selection Criteria: The choice of alkylating agent depends on the downstream application. NEM is the fastest alkylator, ideal for highly unstable modifications. Iodoacetamide and its derivative iodoacetic acid are preferred for mass spectrometry (MS)-based workflows, as they yield a predictable mass addition.
2. Critical Experimental Parameters:
- pH: Alkylation is most efficient at pH 8.0-8.5, where the thiolate anion (-S⁻) is predominant.
- Concentration & Time: A large molar excess (e.g., 20-50 mM) over total thiols is required for complete alkylation within 10-30 minutes in the dark.
- Temperature: Room temperature or 37°C is standard; 4°C may be used for extremely labile modifications but requires longer incubation.
- Quenching: Excess alkylating agent must be quenched (e.g., with DTT or excess cysteine) before downstream steps like tryptic digestion.
3. Quantitative Comparison of Common Alkylating Agents: Table 1: Properties and Applications of Key Alkylating Agents
| Reagent | Mechanism | Speed | Mass Addition (Da) | MS Compatibility | Primary Use Case |
|---|---|---|---|---|---|
| NEM | Michael addition | Very Fast | +125.12 | Good, but can undergo hydrolysis | Rapid trapping in kinetic studies, activity assays |
| Iodoacetamide (IAA) | Nucleophilic substitution | Moderate | +57.02 | Excellent; stable adduct | Standard proteomics, 2D gels |
| Iodoacetic Acid (IAA-COOH) | Nucleophilic substitution | Moderate | +58.01 (for -COOH) | Excellent; adds negative charge | Proteomics, improves peptide retention in negative mode |
4. Differential Alkylation for Oxidation Mapping: A central protocol in redox proteomics involves sequential alkylation. Reduced thiols are first blocked with a light alkylating agent (e.g., NEM or light IAA). Following reduction of oxidized modifications (disulfides, sulfenic acids), the newly revealed thiols are labeled with a heavy isotope version of the reagent (e.g., ¹³C₂-IAA). The mass shift in MS allows for precise quantification of the oxidation state of individual cysteines.
Detailed Protocols
Protocol 1: Rapid Tissue/Cell Lysis with Concurrent Alkylation for Redox Proteomics
Objective: To instantly trap the native redox state of cysteines during cell disruption. Materials: See "Scientist's Toolkit" below. Procedure:
- Prepare Alkylation Buffer: Freshly prepare ice-cold lysis buffer (e.g., 50 mM HEPES, 150 mM NaCl, 1% NP-40, pH 8.0) supplemented with 20-50 mM NEM or IAA and protease/phosphatase inhibitors. Keep in the dark.
- Lysis: Immediately add cold alkylation buffer to cell pellet or flash-frozen tissue powder (e.g., 1 mL per 10⁷ cells). Vortex vigorously.
- Incubate: Sonicate on ice (if needed for complete lysis) and then incubate the lysate in the dark at room temperature for 30 minutes with gentle agitation.
- Quench & Clear: Add DTT to a final concentration of 10 mM to quench excess alkylating agent. Incubate 15 min. Centrifuge at 16,000 x g for 15 min at 4°C to clear debris.
- Protein Clean-up: Precipitate protein using cold acetone/methanol/chloroform method or proceed to buffer exchange via desalting column into digestion-compatible buffer.
Protocol 2: Sequential Differential Alkylation for Cysteine Oxidation Quantification (ICAT-like)
Objective: To distinguish between reduced and reversibly oxidized cysteine residues. Materials: Light IAA (or NEM), heavy IAA (¹³C₂-D₂-IAA, +61.05 Da), reducing agent (TCEP or DTT). Procedure:
- Initial Alkylation (Block Free Thiols): Take reduced, cleared protein lysate. Adjust to pH 8.0 with Tris-HCl. Add light IAA to 20 mM final. Incubate 30 min, in dark, RT. Quench with 10 mM DTT for 15 min.
- Reduction of Oxidized Species: Add a potent reducing agent like TCEP to 10 mM final. Incubate 30 min at 37°C to reduce all reversibly oxidized cysteines (disulfides, S-nitrosothiols, sulfenic acids).
- Secondary Alkylation (Label Newly Reduced Thiols): Add heavy IAA (¹³C₂-D₂-IAA) to 40 mM final. Incubate 30 min, in dark, RT.
- Digestion & MS Analysis: Quench reaction, digest with trypsin, and analyze by LC-MS/MS. The paired light/heavy peptide signals allow quantification of the oxidation fraction at each cysteine site.
Visualizations
Diagram Title: Sequential Differential Alkylation Workflow
Diagram Title: IAA Alkylation Chemical Mechanism
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions for Redox Trapping
| Reagent / Material | Function & Purpose | Key Considerations |
|---|---|---|
| N-Ethylmaleimide (NEM) | Fast, irreversible thiol alkylator. Ideal for kinetic trapping and activity assays. | Light-sensitive. Prepare fresh in ethanol/DMSO. Quench with excess thiol. |
| Iodoacetamide (IAA) | Standard alkylator for MS proteomics. Adds a consistent +57.0215 Da mass tag. | Light- and air-sensitive. Use in dark, at pH ~8.0. Can alkylate amines if over-incubated. |
| ¹³C/¹⁵N-Deuterated IAA | Isotope-coded heavy alkylator for quantitative redox proteomics (e.g., +61.05 Da). | Enables precise MS-based quantification of oxidized vs. reduced cysteine pairs. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Strong, non-thiol reducing agent. Reduces all reversible cysteine oxidations. | More stable than DTT, works at acidic pH. Does not interfere with alkylation. |
| HEPES or Tris Buffer, pH 8.0-8.5 | Alkylation reaction buffer. Maintains optimal pH for thiolate formation. | Avoid amine-containing buffers (e.g., glycine) that can compete with alkylation. |
| Protease/Phosphatase Inhibitor Cocktails | Preserves protein integrity and phosphorylation state during lysis/alkylation. | Use EDTA-free versions if studying metal-dependent oxidation (e.g., by Cu/Zn). |
| Mass Spectrometry-Compatible Detergent (e.g., SDC) | Facilitates protein solubilization for downstream digestion and LC-MS/MS. | Must be acid-cleavable to avoid interference during MS analysis. |
The Biotin-Switch Technique (BST) and Modified S-Nitrosylation (SNO) Protocols.
This document details the Biotin-Switch Technique (BST) and its modern derivations, a cornerstone methodology for the specific detection of S-nitrosylated proteins (SNO-proteins). Within the broader thesis on "Methods for assessing cysteine oxidation states in proteins," the BST addresses a critical niche. Cysteine residues can undergo diverse oxidative post-translational modifications (PTMs) including disulfide bond formation, sulfenation (-SOH), sulfination (-SO2H), and S-glutathionylation. S-nitrosylation, the reversible addition of a nitric oxide (NO) group to a cysteine thiol to form an S-nitrosothiol (SNO), is a key redox-based signaling mechanism. The BST specifically identifies this PTM by exploiting the labile S-NO bond, allowing it to be distinguished from other, more stable cysteine oxidations. This specificity is paramount for elucidating the role of NO signaling in physiological and pathological processes, from cardiovascular function to neurodegeneration and cancer, making it highly relevant for drug development targeting redox pathways.
Core Principles and Evolution of the BST
The classical BST, developed by Jaffrey and Snyder (2001), involves three sequential chemical steps designed to convert labile SNO modifications into a stable, detectable biotin tag:
- Blocking of Free Thiols: All unmodified (reduced) cysteine thiols are covalently and irreversibly blocked with methyl methanethiosulfonate (MMTS).
- Selective Reduction of S-NO Bonds: S-nitrosothiols are selectively reduced using ascorbate (or related compounds) to generate new free thiols. This step is highly specific; ascorbate does not reduce disulfides, sulfenic acids, or metal-bound thiols under optimized conditions.
- Biotinylation and Detection: The newly revealed thiols are labeled with a sulfhydryl-specific biotinylating agent (e.g., Biotin-HPDP). The biotinylated proteins can then be affinity-purified on streptavidin beads and detected by immunoblotting or identified by mass spectrometry.
Modern "Modified BST" protocols address key limitations of the original method:
- Artifact Control: Use of metal chelators (e.g., EDTA, neocuproine) to prevent Cu⁺-mediated ascorbate reduction of disulfides and trans-nitrosylation.
- Improved Specificity: Substitution of ascorbate with alternative reducing agents like copper(II) sulfate with ascorbate (CuAsc) or trialkylphosphines for more controlled reduction.
- Enhanced Sensitivity: Development of resin-assisted capture (RASC) methods, where thiols are captured directly onto solid supports after reduction, reducing background and improving compatibility with MS.
Comparative Table of BST Methodologies
Table 1: Comparison of BST Variants and Key Reagents
| Method Variant | Key Reduction Agent | Primary Advantage | Key Consideration / Limitation | Typical Application |
|---|---|---|---|---|
| Classical BST (Jaffrey & Snyder) | Sodium Ascorbate | Simplicity, foundational protocol. | Risk of ascorbate-mediated reduction of disulfides (artifacts); metal-dependent. | Initial detection of SNO-proteins in cell lysates. |
| "Switch" Method (Forrester et al.) | Cu(II) + Ascorbate (CuAsc) | Increased sensitivity and speed of reduction. | Requires precise optimization of Cu(II) concentration to avoid non-specific reduction. | Detecting low-abundance SNO-proteins. |
| Trialkylphosphine-Based | Triethylphosphine (TEP) | Metal-independent; high selectivity for SNO vs. disulfides. | Requires anaerobic conditions; TEP is toxic and volatile. | Studies where metal chelation is problematic. |
| Resin-Assisted Capture (RASC) | Ascorbate or CuAsc | Reduces background; proteins captured directly on thiol-reactive resin. | Multi-step washing on resin; may lose some proteins. | Proteomic identification of S-nitrosylated sites (MS). |
| SNO-RAC | Ascorbate | Direct capture eliminates elution step; compatible with non-denaturing conditions. | Requires careful control of blocking efficiency. | Analysis of SNO-protein complexes. |
Detailed Experimental Protocols
Protocol A: Standard BST for Immunoblot Detection
Objective: To detect specific S-nitrosylated proteins in cell or tissue lysates.
I. Solutions and Reagents (Prepare fresh daily):
- HENS Buffer: 250 mM HEPES-NaOH (pH 7.7), 1 mM EDTA, 0.1 mM neocuproine, 2.5% SDS.
- Blocking Buffer: HENS buffer with 20 mM Methyl Methanethiosulfonate (MMTS).
- Reduction Buffer: HENS buffer with 20 mM Sodium Ascorbate.
- Biotinylation Buffer: HENS buffer with 0.4 mM Biotin-HPDP (from 4 mM stock in DMSO) and 2.5 mM EDTA (final).
- NeutrAvidin/Agarose Beads
II. Procedure:
- Lysate Preparation: Lyse cells/tissue in HENS buffer (+ protease inhibitors). Clarify by centrifugation (14,000 x g, 15 min, 4°C). Determine protein concentration. Use >1 mg total protein for reliable detection.
- Free Thiol Blocking: To 100 µL of lysate (1 mg/mL), add 4 volumes (400 µL) of Blocking Buffer. Incubate at 50°C for 20 min with frequent vortexing.
- Acetone Precipitation: Add 2 volumes of pre-chilled acetone. Incubate at -20°C for 20 min. Pellet protein (8,000 x g, 10 min, 4°C). Wash pellet 2x with 70% acetone. Air-dry pellet briefly.
- Selective Reduction of SNO: Resuspend pellet in 100 µL Reduction Buffer. Incubate at room temperature for 1 hour.
- Biotinylation: Add 10 µL of 40 mM Biotin-HPDP stock (in DMSO) to achieve 4 mM final. Incubate at room temperature for 1 hour in the dark.
- Clean-up & Detection: Precipitate proteins with acetone as in Step 3. Resuspend pellet in non-reducing Laemmli buffer.
- Direct Western: Analyze 10-20% of sample by SDS-PAGE/immunoblot with streptavidin-HRP to detect total biotinylated (SNO) proteins.
- Pull-down/Western: Resuspend the remaining sample in neutralization buffer, incubate with NeutrAvidin beads (1 hr), wash extensively, elute with Laemmli buffer + β-mercaptoethanol, and probe for your protein of interest by immunoblot.
Protocol B: SNO-RAC (Resin-Assisted Capture) for Proteomics
Objective: To enrich and identify the S-nitrosylated proteome by mass spectrometry.
I. Key Materials:
- Thiopropyl Sepharose 6B resin
- Elution Buffer: 20 mM HEPES (pH 7.7), 100 mM NaCl, 1 mM EDTA, 20 mM β-mercaptoethanol (or 100 mM DTT).
II. Procedure:
- Blocking: Perform as in Protocol A, Steps 1-3.
- Resin Preparation: Wash 50 µL packed Thiopropyl Sepharose beads 3x with HENS buffer.
- Capture: Resuspend acetone-precipitated protein pellet from Step 1 in HENS buffer + 20 mM Sodium Ascorbate. Immediately incubate with the prepared resin for 30 min at room temperature with rotation. The ascorbate reduces SNO bonds, and the newly freed thiols couple to the resin via disulfide exchange.
- Washing: Wash beads sequentially with:
- Wash 1: HENS buffer (2x)
- Wash 2: 2 M NaCl in HENS (2x)
- Wash 3: 10% Isopropanol in HENS (1x)
- Wash 4: 25 mM NH₄HCO₃, pH 8.0 (2x, for MS compatibility)
- Elution: Elute bound proteins with Elution Buffer for 10 min at 37°C. Repeat elution once. Combine eluates.
- Analysis: The eluate can be analyzed by SDS-PAGE/Coomassie or silver stain, or processed for mass spectrometry (trypsin digestion, LC-MS/MS).
Signaling Pathway and Experimental Workflow Diagrams
BST Core Chemical Workflow
NO Signaling via S-Nitrosylation
The Scientist's Toolkit: Essential Reagents & Solutions
Table 2: Key Research Reagent Solutions for BST/SNO Studies
| Reagent / Material | Function / Role in BST | Critical Notes |
|---|---|---|
| Methyl Methanethiosulfonate (MMTS) | Thiol-blocking agent. Methylates free cysteine thiols to prevent non-specific labeling. Small size ensures access to buried residues. | Preferred over N-ethylmaleimide (NEM) for BST due to size and reversibility under strong reducing conditions. |
| Sodium Ascorbate | Selective reducing agent. Specifically reduces S-NO bonds to liberate free thiols without reducing disulfides (under optimized, chelated conditions). | Must be prepared fresh. Effectiveness is Cu⁺-dependent, necessitating chelators to control specificity. |
| Biotin-HPDP | Biotinylating agent. Contains a disulfide bond that reacts with the newly freed thiols, introducing a biotin tag for affinity capture/detection. | The HPDP linker allows elution with reducing agents (β-Me, DTT). Light-sensitive. |
| Neocuproine | Specific Cu⁺ chelator. Inhibits Cu⁺-mediated ascorbate reduction of disulfides, a major source of artifact in BST. | Crucial for specificity. More effective than EDTA for this specific chelation. |
| Thiopropyl Sepharose 6B | Activated thiol resin. Used in RASC methods. Captures proteins via disulfide exchange after SNO reduction, enabling direct on-resin washing. | Eliminates the need for biotinylation and streptavidin beads, reducing non-specific binding. |
| Triethylphosphine (TEP) | Metal-independent reducing agent. Directly reduces SNO bonds without metal catalysis, offering an alternative pathway. | Used under strict anaerobic conditions. Offers high selectivity but requires specialized handling. |
| Streptavidin/NeutrAvidin Beads | Affinity matrix. Binds biotinylated proteins with high affinity for purification and concentration prior to analysis. | NeutrAvidin (deglycosylated) has lower non-specific binding than native streptavidin. |
Application Notes
Within the broader thesis on methods for assessing cysteine oxidation states in proteins, the detection of sulfenic acid (-SOH) is a critical challenge due to its transient nature and role as a key redox signaling intermediate. Dimedone (5,5-dimethyl-1,3-cyclohexanedione) and its derivatives serve as specific, nucleophilic probes that form stable thioether adducts with sulfenic acids, enabling detection, quantification, and identification of these modifications.
Key Advantages:
- Specificity: Minimal reactivity with other cysteine oxidations (e.g., disulfides, sulfinic acid) or reduced thiols under physiological conditions.
- Versatility: Functionalized derivatives allow for "click chemistry" conjugation, biotinylation, or fluorophore tagging for diverse detection strategies (gel-based, mass spectrometry, microscopy).
- Cell Permeability: Many probes (e.g., DAz-2, DYn-2) are cell-permeable, enabling live-cell imaging and labeling.
Primary Applications:
- Biomarker Discovery: Profiling proteins sensitive to oxidative stress in disease models.
- Pathway Analysis: Mapping redox-dependent signaling networks in cells.
- Drug Mechanism Studies: Identifying target engagement and off-target effects of redox-modulating therapeutics.
Experimental Protocols
Protocol 1: In-Gel Fluorescence Detection of Protein Sulfenylation in Cell Lysates
Objective: To label and detect sulfenylated proteins from mammalian cell lysates using a fluorescent dimedone derivative.
Reagents & Materials: See "Research Reagent Solutions" table.
Procedure:
- Cell Treatment & Lysis:
- Culture and treat cells (e.g., with H₂O₂) in a 6-well plate.
- Aspirate media, wash with PBS, and lyse cells on ice for 10 min using 200 µL of modified RIPA lysis buffer (containing 20 mM NEM to block free thiols and prevent post-lysis artifacts).
- Clarify lysate by centrifugation (16,000 × g, 15 min, 4°C).
- Determine protein concentration (e.g., BCA assay).
Sulfenic Acid Labeling:
- Prepare a fresh 50 mM stock of DCP-Bio1 in DMSO.
- To 50 µg of clarified lysate, add DCP-Bio1 to a final concentration of 100 µM. Incubate for 1 hour at room temperature, protected from light.
"Click Chemistry" Conjugation (if using alkyne/azide probes):
- Note: For direct fluorescent probes like DCP-Rho1, skip to Step 4.
- To the labeled lysate, add (final concentrations):
- CuSO₄: 1 mM
- THPTA Ligand: 1 mM
- Aminoguanidine: 5 mM
- Sodium Ascorbate: 5 mM
- Fluorescent Azide (e.g., Azide-Fluor 488): 50 µM
- Vortex and incubate for 1 hour at room temperature, protected from light.
Detection:
- Terminate the reaction by adding SDS-PAGE loading buffer (non-reducing, without DTT or β-mercaptoethanol).
- Resolve proteins by SDS-PAGE (4-20% gradient gel recommended).
- Visualize fluorescently labeled proteins using a gel scanner (e.g., Typhoon) with appropriate laser/filter settings (e.g., 488 nm excitation/520 nm emission for Azide-Fluor 488).
- For total protein loading control, stain gel with Coomassie or SYPRO Ruby.
Protocol 2: Chemoproteomic Enrichment and Identification of Sulfenylated Proteins
Objective: To enrich and identify sulfenylated peptides via mass spectrometry using a biotinylated dimedone probe.
Procedure:
- Labeling & Digestion:
- Label 1-2 mg of cell lysate protein with 200 µM biotin-conjugated dimedone probe (e.g., DCP-Bio1) for 2 hours at room temperature.
- Precipitate proteins with cold acetone, resuspend in digestion buffer (8 M urea, 100 mM Tris, pH 8.0).
- Reduce with 5 mM DTT (30 min, 37°C), alkylate with 15 mM iodoacetamide (30 min, RT in dark), then quench with 5 mM DTT.
- Dilute urea to < 2 M with 100 mM Tris, pH 8.0. Digest with trypsin/Lys-C mix (1:50 w/w) overnight at 37°C.
- Acidify with formic acid (FA) to pH ~3, desalt with C18 Sep-Pak cartridge, and dry.
Streptavidin Enrichment:
- Reconstitute peptides in PBS with 0.2% SDS.
- Pre-clear with control agarose beads for 1 hour at 4°C.
- Incubate supernatant with high-capacity streptavidin-agarose beads overnight at 4°C with rotation.
Washing & Elution:
- Wash beads sequentially with: (1) PBS + 0.2% SDS, (2) PBS, (3) 50 mM ammonium bicarbonate.
- Elute bound peptides with 70% acetonitrile, 1% FA, or with 2.5 mM biotin in PBS. Dry eluate.
LC-MS/MS Analysis:
- Reconstitute in 0.1% FA and analyze by nano-LC-MS/MS on a high-resolution instrument.
- Database Search Parameters: Include variable modifications: +136.0528 Da on Cys (for dimedone adduct), +57.0215 Da on Cys (carbamidomethylation), +15.9949 Da on Met (oxidation). Set carbamidomethylation as fixed.
Data Tables
Table 1: Common Dimedone-Based Chemical Probes
| Probe Name | Reactive Group | Tag/Handle | Key Feature | Typical Working Concentration |
|---|---|---|---|---|
| DCP-Bio1 | Dimedone | Biotin | Affinity enrichment; Western blot | 50-200 µM |
| DAz-2 | Dimedone | Azide | "Click" to alkyne-fluor/biotin; cell-permeable | 100-500 µM |
| DYn-2 | Dimedone | Alkyne | "Click" to azide-fluor/biotin; cell-permeable | 100-500 µM |
| DCP-Rho1 | Dimedone | Rhodamine | Direct fluorescence; microscopy & gels | 10-50 µM |
| β-Estradiol-Dimedone | Dimedone | β-Estradiol | Targets estrogen receptor contexts | 1-10 µM |
Table 2: Representative Quantitative Data from Sulfenylation Studies
| Study Model | Probe Used | Key Finding (Quantified) | Detection Method |
|---|---|---|---|
| A431 Epidermal Cells (EGF Stimulation) | DAz-2 | ~250 proteins labeled; PTP1B labeling increased 4.2-fold post-EGF | Chemoproteomics, WB |
| Cardiac Myocytes (H₂O₂ Stress) | DCP-Bio1 | 15% increase in global sulfenylation at 50 µM H₂O₂ vs. control | In-gel fluorescence |
| Liver Tissue (Aged vs. Young Mice) | DYn-2 | 32% more sulfenylated proteins in aged mitochondrial fractions | Chemoproteomics |
Visualizations
Title: Specific Trapping of Transient Sulfenic Acid by Dimedone Probes
Title: Workflow for Detecting Sulfenylated Proteins
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function & Role in Experiment |
|---|---|
| DCP-Bio1 (Biotin-Conjugated Dimedone) | The core reagent. Biotin handle allows streptavidin-based enrichment for Western blot or mass spectrometry. |
| DAz-2 or DYn-2 (Azide/Alkyne Probes) | Cell-permeable probes for live-cell labeling. Enable flexible conjugation via copper-catalyzed azide-alkyne cycloaddition (CuAAC). |
| N-Ethylmaleimide (NEM) | Thiol alkylating agent. Used to block free cysteine thiols post-lysis to prevent post-harvest oxidation and artifact formation. |
| High-Capacity Streptavidin Agarose | For efficient capture of biotinylated proteins/peptides prior to elution and analysis by MS or blot. |
| Triazoleligand (e.g., THPTA) | Copper chelator for CuAAC "click" reactions. Enhances reaction efficiency and reduces copper-induced protein/peptide degradation. |
| Modified RIPA Lysis Buffer | Must be supplemented with NEM (20-50 mM) and sometimes metal chelators (EDTA) to preserve the native sulfenylation state during extraction. |
| Anti-Biotin Antibody (HRP Conjugated) | For direct Western blot detection of biotinylated dimedone adducts without the need for a "click" step. |
| Non-Reducing SDS-PAGE Buffer | Critical. Must omit DTT or β-mercaptoethanol to avoid reduction and cleavage of the dimedone-thioether adduct. |
Thesis Context: This document details application notes and protocols for the identification and quantification of protein cysteine oxidation, a critical post-translational modification in redox signaling, stress response, and disease. These methods are core components of a broader thesis focused on developing robust, sensitive, and comprehensive Methods for assessing cysteine oxidation states in proteins.
Application Notes
Cysteine oxidation, from reversible modifications like S-sulfenylation (-SOH), S-glutathionylation (-SSG), and disulfide formation (-S-S-) to irreversible oxidations, regulates protein function and cellular signaling. Bottom-up proteomics coupled with selective enrichment is the gold standard for system-wide profiling. Key challenges include the labile nature of some modifications, sub-stoichiometric abundance, and the need to preserve oxidation states during sample preparation.
Table 1: Comparison of Major Oxidized Cysteine Enrichment Strategies
| Enrichment Method | Target Modification(s) | Principle | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Biotin Switch Technique (BST) & Derivatives | S-Nitrosylation (-SNO), S-Sulfenylation (-SOH) | Selective reduction of target PTM, then biotinylation of nascent thiols. | High specificity for targeted PTM. | Multi-step; risk of artifact formation. |
| Diamide-based Enrichment | Reduced thiols (-SH) | Diamide reacts with free thiols, which can be tagged with a cleavable biotin probe. | Excellent coverage of the reduced cysteine proteome (reversome). | Does not directly enrich oxidized forms. |
| Oxidized Cysteine Resin-Assisted Capture (OxRAC) | Disulfides, sulfenic acids | Direct coupling of oxidized cysteines to solid-state thiol-reactive resin. | Simpler workflow; minimizes scrambling. | May miss some acid-labile modifications. |
| Iodoacetyl TMT-based (iodoTMT) | Reduced thiols (-SH) | Alkylation of free thiols with isobaric mass tags for multiplexed quantification. | Enables multiplexed (up to 11-plex) quantification of thiol occupancy. | Requires access to a high-resolution MS; tags can be expensive. |
Table 2: Quantitative Data from a Representative iodoTMT Study of H₂O₂-treated Cells
| Protein (Gene Symbol) | Cysteine Site | Condition 1 (Control) TMT Ratio | Condition 2 (H₂O₂) TMT Ratio | Fold Change (Oxidation) | p-value |
|---|---|---|---|---|---|
| Peroxiredoxin 1 (PRDX1) | Cys52 | 0.12 | 3.45 | 28.8 | 1.2e-08 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Cys152 | 0.98 | 0.15 | 0.15 (Reduction) | 0.003 |
| Actin, cytoplasmic 1 (ACTB) | Cys374 | 1.05 | 2.33 | 2.22 | 0.021 |
| Protein tyrosine phosphatase 1B (PTP1B) | Cys215 | 0.08 | 2.98 | 37.3 | 4.5e-09 |
Experimental Protocols
Protocol 1: Oxidized Peptide Enrichment via Oxidized Cysteine Resin-Assisted Capture (OxRAC)
Objective: To enrich peptides containing cysteines oxidized to sulfenic acid or disulfide bonds.
Reagents & Materials: See "The Scientist's Toolkit" below.
Procedure:
- Cell Lysis under Non-Reducing Conditions: Lyse cells or tissue in OxRAC Lysis Buffer (50 mM Tris-HCl pH 7.5, 1% SDC, 1x protease/phosphatase inhibitors, 50 mM N-ethylmaleimide (NEM), 1 mM EDTA). Incubate 30 min at 50°C with frequent vortexing. NEM alkylates free thiols to block reduction and artifact formation.
- Protein Clean-up & Quantification: Precipitate proteins using the methanol-chloroform method. Wash pellet twice with cold methanol. Air-dry and resuspend in 50 mM Tris, 1% SDC, pH 7.5. Quantify via BCA assay.
- Trypsin Digestion: Dilute protein to 1 mg/mL. Add trypsin at a 1:50 (w/w) enzyme-to-protein ratio. Digest overnight at 37°C.
- Peptide Clean-up (StageTip): Acidify peptides with 1% TFA to a final concentration of 0.5% TFA. Desalt using C18 StageTips. Elute peptides with 60% ACN, 0.1% FA. Dry in a vacuum concentrator.
- OxRAC Enrichment: a. Resuspend dried peptides in Binding/Wash Buffer (50 mM HEPES, 100 mM NaCl, 1% SDC, pH 7.0). b. Wash Thiopropyl Sepharose 6B resin twice with Binding/Wash Buffer. c. Incubate peptide mixture with 30 µL resin slurry for 1 hour at room temperature with end-over-end rotation. d. Pellet resin (800 x g, 2 min) and carefully remove supernatant (contains non-oxidized peptides). e. Wash resin 3x with Binding/Wash Buffer, then 3x with MS-grade water.
- Elution of Oxidized Peptides: a. To elute peptides bound via disulfide linkages, add 100 µL of Elution Buffer (20 mM DTT in Binding/Wash Buffer). Incubate 30 min at RT with mixing. b. Pellet resin and collect supernatant (Eluate 1). c. For peptides bound via sulfenic acids (thiosulfinate bond), add 100 µL of 10 mM Sodium Ascorbate (in water). Incubate 15 min at RT. Pellet and collect supernatant (Eluate 2). d. Combine Eluate 1 and 2.
- Post-Enrichment Clean-up & MS Analysis: Acidify combined eluates with TFA. Desalt using C18 StageTips. Dry peptides and resuspend in 0.1% FA for LC-MS/MS analysis.
Protocol 2: Multiplexed Quantification of Thiol Oxidation using Iodoacetyl TMT (iodoTMT)
Objective: To compare the redox state of cysteine residues across multiple experimental conditions in a single MS run.
Procedure:
- Free Thiol Blocking in Situ: Treat cells under experimental conditions (e.g., control, H₂O₂, drug). Immediately aspirate media and lyse cells in Lysis Buffer (6 M Guanidine HCl, 100 mM Tris, 1 mM EDTA, pH 8.5) containing 50 mM NEM. Sonicate and incubate 1 hour at RT in the dark.
- Protein Clean-up: Precipitate proteins with cold acetone. Wash pellet twice. Resuspend in Dissolution Buffer (6 M Guanidine HCl, 100 mM TEAB, pH 8.5).
- Reduction of Oxidized Cysteines: Reduce reversibly oxidized cysteines by adding Tris(2-carboxyethyl)phosphine (TCEP) to 10 mM. Incubate 1 hour at 55°C.
- IodoTMT Labeling of Newly Reduced Thiols: Add iodoTMT reagent (dissolved in anhydrous DMSO) to a final concentration of 2 mM. Incubate for 1 hour at RT in the dark. Each condition receives a unique isobaric TMT channel (e.g., 126, 127N, 127C, etc.).
- Quenching & Pooling: Quench the reaction with 5 mM DTT for 15 min. Combine equal protein amounts from each TMT-labeled condition into a single tube.
- Trypsin Digestion & Clean-up: Dilute guanidine HCl concentration to <1 M with 100 mM TEAB. Digest with trypsin/Lys-C overnight at 37°C. Desalt peptides via C18 solid-phase extraction.
- Anti-TMT Immunoaffinity Enrichment: Use an anti-TMT antibody resin to enrich labeled peptides, significantly reducing background. Follow manufacturer's instructions for binding, washing, and elution (typically with 0.2% TFA).
- LC-MS/MS Analysis: Analyze enriched peptides on a high-resolution tandem mass spectrometer. Use higher-energy collisional dissociation (HCD) for fragmentation to report TMT reporter ions.
Data Analysis Workflow
Diagram 1: Data Analysis Pipeline for Cysteine Oxidoproteomics
Diagram 2: Generalized Redox Signaling Pathway Involving Cysteine Oxidation
The Scientist's Toolkit
Table 3: Essential Research Reagents and Materials for Oxidized Cysteine Proteomics
| Item | Function / Purpose | Example Product / Note |
|---|---|---|
| N-Ethylmaleimide (NEM) | Alkylating agent that covalently blocks free thiols (-SH) during lysis to prevent post-lysis oxidation/scrambling. | Must be fresh; prepare stock in ethanol or ACN. |
| Iodoacetamide (IAM) | Alternative alkylating agent, often used after reduction in standard proteomics to block all thiols. | Light-sensitive. Typically used after DTT/TCEP reduction. |
| Thiopropyl Sepharose 6B | Resin for OxRAC. Contains pyridyl disulfide groups that form mixed disulfides with reduced thiols. | From Cytiva. Critical for direct enrichment of oxidized peptides. |
| Iodoacetyl TMTpro 16plex | Isobaric mass tags with an iodoacetyl group for multiplexed quantification of reduced cysteine residues. | Thermo Fisher Scientific. Enables 16-plex experiment design. |
| Trypsin/Lys-C Mix, MS Grade | Protease for generating peptides. Lys-C improves digestion efficiency in denaturing buffers. | Promega, Thermo Fisher. Essential for bottom-up workflow. |
| StageTips with C18 Material | Low-cost, in-house micro-spin columns for peptide desalting and clean-up. | Use Empore C18 disks or commercial alternatives. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Reducing agent to reduce disulfide bonds. More stable and effective than DTT in many buffers. | Preferred for iodoTMT protocol. |
| High pH Reversed-Phase Peptide Fractionation Kit | For fractionating complex peptide mixtures pre-MS to increase proteome coverage. | Thermo Fisher, Pierce. Used after enrichment. |
| Anti-TMT Antibody Resin | Immunoaffinity resin for enriching TMT-labeled peptides, dramatically reducing non-labeled background. | Thermo Fisher Scientific. Key for iodoTMT sensitivity. |
| LC-MS/MS System | High-resolution, high-mass-accuracy mass spectrometer coupled to nanoflow UHPLC. | Orbitrap Eclipse, timsTOF, etc. Fundamental analytical tool. |
Activity-Based Protein Profiling (ABPP) and Chemoproteomic Platforms for Cysteine Reactivity Screening
Within the broader thesis on "Methods for assessing cysteine oxidation states in proteins," understanding the functional reactivity of cysteine residues is paramount. Cysteine oxidation states, including sulfenic (-SOH), sulfinic (-SO2H), and sulfonic (-SO3H) acids, or disulfide bonds, directly modulate protein function and cellular signaling. Activity-Based Protein Profiling (ABPP) coupled with chemoproteomics provides a powerful, functional platform to screen and characterize reactive, ligandable, and redox-sensitive cysteines proteome-wide, offering a direct readout on their chemical and oxidation states in native biological systems.
ABPP for cysteine reactivity profiling typically employs electrophilic probes that covalently modify nucleophilic, reduced cysteines. Competition with small molecules or changes in redox state alter labeling efficiency, enabling quantification of cysteine reactivity and oxidation.
Table 1: Common Cysteine-Directed Activity-Based Probes (ABPs)
| Probe Name | Core Reactivity Group | Target Cysteine State | Key Application | Typical Concentration Range |
|---|---|---|---|---|
| Iodoacetamide (IA)-Alkyne | Iodoacetamide | Reduced (Thiolate anion, -S⁻) | Broad, non-selective profiling | 100 – 500 µM |
| N-Ethylmaleimide (NEM)-Alkyne | Maleimide | Reduced (-S⁻) | General reactivity screening | 50 – 200 µM |
| Desthiobiotin-Iodoacetamide (DTB-IA) | Iodoacetamide | Reduced (-S⁻) | Enrichment & quantification | 200 – 1000 µM |
| Nucleophile-Oriented ABPs (e.g., CyNHM) | Cyanoacetamide | Hyper-reactive, potentially oxidized (e.g., sulfenylated) | Detection of transient oxidation states | 10 – 100 µM |
Table 2: Representative Chemoproteomic Studies Quantifying Redox-Sensitive Cysteines
| Study Focus (Year) | Platform Used | # of Cysteines Quantified | # Found Redox-Sensitive (e.g., H2O2-responsive) | Key Quantitative Finding |
|---|---|---|---|---|
| Global profiling of S-sulfenylation (2023) | Desthiobiotin-1,3-cyclopentanedione (CPD) probe + LC-MS/MS | > 1,000 | ~ 400 | ~40% of quantified cysteines showed >2-fold increase in labeling upon H2O2 treatment. |
| Covalent ligand screening (2022) | IsoTOP-ABPP (IA-alkyne with isotopically labeled TEV tags) | ~ 10,000 | N/A | Identified ligands for > 700 cysteines; KD values ranged from 1 nM to 100 µM. |
| Kinase cysteome profiling (2023) | NEM-based quantitative chemoproteomics | ~ 5,000 | ~ 800 | ~16% of kinase-domain cysteines displayed significant reactivity shift under oxidative stress. |
Detailed Experimental Protocols
Protocol 1: IsoTOP-ABPP for Quantifying Cysteine Reactivity and Small-Molecule Engagement
Application: Identifying and quantifying ligandable cysteines and measuring changes in reactivity (e.g., due to oxidation).
Materials:
- Cells or tissue lysate (1-2 mg/mL protein in PBS with protease inhibitors).
- IA-alkyne probe (stock 50 mM in DMSO).
- Test compound or DMSO vehicle.
- Click chemistry reagents: CuSO4, THPTA ligand, sodium ascorbate, Azide-PEG3-Biotin (or Azide-TEV-isoTag for IsoTOP).
- Streptavidin beads.
- On-bead trypsin digestion reagents.
- LC-MS/MS system.
Method:
- Lysate Treatment & Probing: Divide lysate into two aliquots. Pre-treat one with compound (e.g., 10 µM, 30 min, 25°C) and the other with DMSO. Add IA-alkyne probe to a final concentration of 200 µM. Incubate 1 hr, 25°C, in the dark.
- Click Chemistry: Add CuSO4 (100 µM final), THPTA ligand (300 µM final), sodium ascorbate (1 mM final), and Azide-PEG3-Biotin (50 µM final) directly to the labeling reaction. Incubate with rotation for 1 hr at 25°C.
- Protein Precipitation & Clean-up: Precipitate proteins using cold methanol/chloroform. Wash pellet 2x with cold methanol.
- Streptavidin Enrichment: Resuspend protein pellets in PBS with 1% SDS. Dilute SDS to 0.1% with PBS. Incubate with pre-washed streptavidin beads overnight at 4°C with rotation.
- On-Bead Processing: Wash beads sequentially with: 1% SDS in PBS, 6M Urea in PBS, PBS, and water. Perform on-bead trypsin digestion (2 µg trypsin, 37°C, overnight).
- LC-MS/MS Analysis: Desalt peptides and analyze by LC-MS/MS. Identify and quantify peptides. For IsoTOP-ABPP, use isotopically labeled TEV tags to generate ratio (R) values signifying compound-induced probe displacement.
Protocol 2: Direct Detection of Sulfenylated Cysteines Using a CPD Probe
Application: Mapping and quantifying S-sulfenylation, a key oxidative post-translational modification.
Materials:
- Live cells or fresh lysate.
- Dinucleophile probe (e.g., DTB- or BTD-linked 1,3-cyclopentanedione).
- Lysis buffer (PBS with 1% NP-40, protease inhibitors, 50 mM NEM to block free thiols).
- Streptavidin beads, Elution buffer (2 mM biotin in PBS or 2x Laemmli buffer with DTT).
Method:
- Probe Labeling in Live Cells: Treat live cells with oxidant (e.g., H2O2, 100-500 µM, 5 min) or vehicle. Add CPD probe (e.g., CyNHM or BTD-CPD) to culture medium (final 50 µM). Incubate 30-60 min, 37°C.
- Cell Lysis & Blocking: Harvest cells, lyse in NEM-containing buffer to alkylate all unreacted, reduced cysteines.
- Click Chemistry (if using alkyne-functionalized CPD): Perform click reaction with Azide-PEG3-Biotin as in Protocol 1.
- Enrichment & Analysis: Enrich biotinylated proteins on streptavidin beads. Wash stringently. Elute with 2 mM biotin or boil in Laemmli buffer with DTT. Analyze by western blot or process for LC-MS/MS (on-bead digestion as in Protocol 1).
Visualizations
Title: ABPP Workflow for Redox Cysteine Profiling
Title: Cysteine Oxidation States and Probe Reactivity
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Cysteine-Directed ABPP
| Reagent / Material | Function & Rationale | Example Vendor / Cat. No. (Representative) |
|---|---|---|
| IA-Aminoalkyne (Iodoacetamide-Alkyne) | The foundational broad-spectrum cysteine-reactive probe. The alkyne handle enables bioorthogonal click chemistry conjugation. | Thermo Fisher, Click Chemistry Tools |
| Azide-PEG3-Biotin | Azide-containing biotin reagent for CuAAC click chemistry with alkyne-labeled proteins, enabling streptavidin-based enrichment and detection. | Sigma-Aldrich, Click Chemistry Tools |
| CPD-based Probes (e.g., BTD-CPD) | Dinucleophile probes that specifically react with sulfenic acid (-SOH), allowing for direct detection of this transient oxidation state. | MilliporeSigma, Custom synthesis |
| Tetrazine-based Probes | For inverse-electron demand Diels-Alder (IEDDA) click chemistry; faster and copper-free, useful for live-cell labeling. | Click Chemistry Tools, SibTech |
| Tandem Mass Tag (TMT) or Isobaric Tags | Enable multiplexed quantitative proteomics (e.g., 11-plex), allowing comparison of many conditions (redox states, doses, timepoints) in one MS run. | Thermo Fisher Scientific |
| High-Stringency Streptavidin Beads | For efficient capture of biotinylated proteins/peptides with low non-specific binding. Critical for deep proteome coverage. | Pierce Streptavidin Ultralink Resin |
| THPTA Ligand | A copper-chelating ligand for CuAAC click chemistry that protects proteins from copper-induced degradation/oxidation. | Click Chemistry Tools |
| Cell-Permeable Alkylating Agent (e.g., NEM) | Used to rapidly block free thiols during cell lysis, "trapping" the in vivo redox state and preventing post-lysis artifacts. | Sigma-Aldrich |
Within the broader thesis on "Methods for assessing cysteine oxidation states in proteins," this document details a critical experimental approach. Cysteine residues are central redox sensors in biology, with their oxidation states (e.g., sulfenic acid, disulfide, sulfinic acid) dictating protein function in signaling, stress response, and disease. Gel-based assays utilizing differential alkylation and oxidant-dependent mobility shifts offer a foundational, accessible, and semi-quantitative method to detect and differentiate these modifications, particularly suitable for initial characterization and time-course studies.
Theoretical Basis & Application Notes
Principle of Differential Alkylation
The core principle exploits the differential reactivity of reduced versus oxidized cysteine thiols with alkylating agents. Reduced (free thiol) cysteines are blocked with a conventional alkylating agent like N-ethylmaleimide (NEM) or iodoacetamide (IAM) under denaturing but non-reducing conditions. Following reduction with DTT or TCEP, newly exposed thiols (originally oxidized) are then labeled with a distinct alkylating agent, often a maleimide conjugated to a mass tag or fluorophore (e.g., maleimide-PEG or maleimide-biotin). This creates a differential tag signature detectable by gel shift or blotting.
Principle of Oxidant-Dependent Mobility Shifts
Certain cysteine oxidations, particularly to sulfenic acid or intra/intermolecular disulfides, can alter protein conformation and stability under non-reducing conditions, leading to altered electrophoretic mobility in SDS-PAGE. Sulfenic acid formation or disulfide bonding can cause slower migration (band upshift) due to incomplete unfolding, while sometimes causing faster migration. Treatment with reducing agents reverses these shifts, confirming the redox nature of the change.
Table 1: Common Cysteine Modifications and Detectability by Gel-Based Assays
| Modification (State) | Chemical Formula | Reactivity with Alkylator (Pre-Reduction) | Gel Mobility Shift (Non-Reducing) | Detectable by Differential Alkylation? |
|---|---|---|---|---|
| Free Thiol (Reduced) | -SH | High | Baseline | Yes (direct label) |
| Disulfide (S-S) | -S-S- (intra/inter) | None | Often Upshift | Yes (post-reduction label) |
| Sulfenic Acid | -SOH | Low (can be trapped) | Possible Upshift | Yes (with specific traps like dimedone) |
| Sulfinic Acid | -SO₂H | None | Possible Shift | Indirectly |
| Sulfonic Acid | -SO₃H | None | Possible Shift | No |
| S-Nitrosothiol | -SNO | None (labile) | Possible Shift | Yes (via Ascorbate/SNO-specific reagents) |
| S-Glutathionylation | -SSG | None | Possible Upshift | Yes (post-reduction label) |
Key Applications in Research & Drug Development
- Mapping Redox-Sensitive Cysteines: Identifying sensor cysteines in kinases, phosphatases, and transcription factors (e.g., NF-κB, HIF-1α).
- Mechanism of Action Studies: For redox-cycling drugs (e.g., chemotherapeutics like arsenic trioxide) or NRF2 activators.
- Biomarker Discovery: Assessing global or specific protein oxidation in disease models (neurodegeneration, cardiovascular disease).
- Stability & Formulation: Monitoring unwanted oxidation of cysteine-containing biotherapeutics during production and storage.
Detailed Experimental Protocols
Protocol A: Differential Alkylation with Maleimide-PEG Shift Assay
Objective: To detect and semi-quantify reversible cysteine oxidation via a mass-tag gel shift.
Materials:
- Cell lysate or purified protein sample.
- Lysis Buffer: 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, plus fresh 20-50 mM N-ethylmaleimide (NEM) or 50 mM Iodoacetamide (IAM). Add protease inhibitors.
- Wash Buffer: Lysis buffer without detergent.
- Reducing Agent: 1M Dithiothreitol (DTT) or 500 mM Tris(2-carboxyethyl)phosphine (TCEP).
- Alkylating-Tag: 10 mM Maleimide-PEG (e.g., 5kDa or 10kDa, from commercial sources) in PBS or water.
- Quenching Solution: 100 mM DTT.
- 4x Non-Reducing Laemmli Sample Buffer.
- SDS-PAGE system, Western blot equipment.
Procedure:
- Initial Alkylation (Block Free Thiols): Lyse cells/tissue directly in ice-cold NEM/IAM-containing lysis buffer. Incubate on ice for 30 min with occasional vortexing. This alkylates all pre-existing free thiols.
- Protein Cleanup: To remove excess NEM/IAM, precipitate protein using TCA/Acetone or use a desalting spin column equilibrated with Wash Buffer.
- Reduction of Oxidized Thiols: Resuspend or dilute the protein sample. Add DTT or TCEP to a final concentration of 10-20 mM. Incubate at 37-50°C for 30 min. This reduces disulfides and other reversible oxidations.
- Differential Alkylation (Tag Ex-Oxidized Thiols): Add Maleimide-PEG to a final concentration of 1-2 mM. Incubate at room temperature in the dark for 1-2 hours. This labels thiols that were originally oxidized.
- Quench Reaction: Add excess DTT (to 50 mM final) to quench unreacted maleimide-PEG. Incubate 10 min.
- Analysis: Add Non-Reducing Sample Buffer (do not add DTT/β-Me). Boil samples for 5 min. Run SDS-PAGE. A higher molecular weight band shift indicates PEGylation of proteins containing reversibly oxidized cysteines. Confirm by Western blot.
Protocol B: Oxidant-Dependent Mobility Shift Assay (Non-Reducing vs. Reducing PAGE)
Objective: To visualize conformational changes due to cysteine oxidation via electrophoretic mobility shifts.
Materials:
- Treated/untreated samples.
- Non-Reducing Lysis Buffer: As in Protocol A but OMIT any alkylating agent (NEM/IAM) and reducing agents. May include alkylators only if immediately trapping a specific state.
- Reducing Lysis Buffer: Non-Reducing Lysis Buffer + 20 mM DTT.
- 4x Non-Reducing and 4x Reducing Laemmli Sample Buffer.
- SDS-PAGE system.
Procedure:
- Parallel Sample Preparation:
- Tube A (Non-Reducing Condition): Lyse sample in ice-cold Non-Reducing Lysis Buffer. Immediately mix with 4x Non-Reducing Sample Buffer. Do not boil or boil for <2 min to avoid heat-induced reduction.
- Tube B (Reducing Condition): Lyse sample in ice-cold Reducing Lysis Buffer. Incubate 10 min at 37°C. Mix with 4x Reducing Sample Buffer. Boil as usual.
- Gel Electrophoresis: Load both samples from the same treatment condition on adjacent lanes of the same SDS-PAGE gel.
- Analysis: Compare migration. A band present in the non-reducing lane that runs at a different apparent MW than the band in the reducing lane indicates an oxidation-induced conformational change (e.g., disulfide bond). The reducing lane shows the fully reduced, linearized form.
Data Presentation
Table 2: Example Quantitative Data from a Differential Alkylation-PEG Shift Experiment on Hypothetical Protein Kinase C (PKC)
| Treatment Condition | % Band Shift (PEGylated) | Apparent MW Non-Red. (kDa) | Apparent MW Red. (kDa) | Inferred Redox State Change |
|---|---|---|---|---|
| Control (Untreated) | 15 ± 3% | 80 | 78 | Baseline oxidation |
| H₂O₂ (200 µM, 5 min) | 65 ± 8% | 83 (diffuse) | 78 | Increased reversible oxidation (SOH/SS) |
| H₂O₂ + DTT Rescue | 18 ± 4% | 80 | 78 | Reversal confirms reducible oxidation |
| Diamide (2 mM, 10 min) | 85 ± 5% | 85 | 78 | Extensive disulfide formation |
| NAC Pre-treatment | 10 ± 2% | 80 | 78 | Antioxidant protection |
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions
| Reagent | Function & Critical Notes |
|---|---|
| N-Ethylmaleimide (NEM) | Thiol-alkylating agent. Used to irreversibly block free thiols. Must be fresh; light-sensitive. |
| Iodoacetamide (IAM) | Alternative alkylating agent. Can be used for alkylation before MS, but may be less efficient than NEM in some contexts. |
| Maleimide-PEG (e.g., 5kDa) | Mass-tag alkylator. Creates a clear gel shift upon labeling. Larger PEGs give more pronounced shifts. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Reducing agent. Advantages: more stable than DTT, works at a wider pH range, does not form mixed disulfides. |
| Dithiothreitol (DTT) | Common reducing agent. Breaks disulfide bonds. Unstable in buffer stocks. |
| Dimedone (5,5-dimethyl-1,3-cyclohexanedione) | Specific nucleophilic trap for sulfenic acid (-SOH). Can be conjugated to biotin or probes for detection. |
| Methyl methanethiosulfonate (MMTS) | A thiol-blocking agent that modifies thiols to a form reducible by DTT, useful in some differential protocols. |
| Non-Reducing Sample Buffer | SDS-PAGE buffer lacking β-mercaptoethanol or DTT. Critical for preserving oxidation states during sample prep. |
| Protease Inhibitor Cocktail (without EDTA where relevant) | Prevents protein degradation. EDTA can chelate metals and affect some metal-catalyzed oxidations. |
| Sodium Arsenite / IAA (for specific trapping) | Used in sequential alkylation protocols to differentiate between sulfenic acid and other modifications. |
Visualization Diagrams
Diagram 1 Title: Differential Alkylation Workflow for Redox Proteomics
Diagram 2 Title: Decision Logic for Interpreting Gel Mobility Shifts
Overcoming Experimental Hurdles: Best Practices for Accurate and Reproducible Redox Analysis
Within the broader thesis on Methods for assessing cysteine oxidation states in proteins, a foundational and non-negotiable pillar is the prevention of artifactual oxidation or reduction during sample preparation. The redox state of catalytic or allosteric cysteine residues is a critical post-translational modification regulating protein function, signaling, and disease. Any deviation from the in vivo state introduced during handling compromises data integrity, leading to erroneous biological conclusions and flawed drug development pipelines. This Application Note details the protocols and principles essential for preserving native cysteine redox states through rigorous sample handling, anaerobic techniques, and rapid quenching.
Table 1: Sources of Artifactual Cysteine Modification During Sample Handling
| Artifact Source | Primary Effect | Typical Consequence | Reported False Oxidation Increase* |
|---|---|---|---|
| Atmospheric Oxygen (O₂) | Non-physiological disulfide formation, sulfenic acid (-SOH) formation. | Overestimation of oxidized species. | 20-50% in non-quenched cell lysates. |
| Lysis Buffer Oxidation | Reaction with dissolved O₂ or metal ions (Fe³⁺, Cu²⁺). | Global background oxidation. | Variable; up to 30% with chelator-free buffers. |
| Sample Heating (>4°C) | Accelerated thiol oxidation. | Loss of reduced thiol signal. | 2-5% per minute at room temp. |
| Alkaline pH (>8.0) during lysis | Deprotonation of thiolate anion (-S⁻), increasing reactivity. | Exaggerated, non-specific alkylation. | Highly variable; pH 8.5 vs 7.4 can double alkylation rate. |
| Delayed Quenching | Continued enzymatic activity (e.g., peroxidases, oxidoreductases). | Dynamic shifts post-homogenization. | Can completely reverse or obscure true state. |
*Compiled from recent literature on redox proteomics. Actual values are system-dependent.
Table 2: Efficacy of Common Quenching/Stabilization Agents
| Agent | Final Concentration | Target | Mechanism | Considerations |
|---|---|---|---|---|
| N-Ethylmaleimide (NEM) | 10-50 mM | Free thiols (-SH) | Irreversible alkylation. | Can be bulky; may inhibit downstream enzymes for activity assays. |
| Iodoacetamide (IAM) | 20-100 mM | Free thiols (-SH) | Irreversible alkylation. | Slower than NEM; light-sensitive. |
| Trichloroacetic Acid (TCA) | 10-20% (w/v) | Proteins / Enzymes | Rapid acid denaturation and precipitation. | Harsh; may cause protein aggregation and co-precipitation of other biomolecules. |
| Methanol / Acetone (Cold) | 80-90% v/v | Proteins / Enzymes | Rapid dehydration and denaturation. | Can be incomplete for some membrane proteins. |
| Perchloric Acid | 5-10% (v/v) | Metabolism / Enzymes | Strong acid denaturation. | Requires careful handling and neutralization. |
Experimental Protocols
Protocol 1: Rapid, Anaerobic Quenching and Lysis for Cultured Mammalian Cells Objective: To instantly arrest metabolism and fix redox states under oxygen-free conditions. Materials: Pre-chilled anaerobic lysis buffer (see Toolkit), sealed degassing system, glove bag/chamber purged with N₂ or Ar, rapid filtration/vacuum quench setup.
- Preparation: Inside an anaerobic chamber (<1 ppm O₂), prepare deaerated lysis buffer containing 50 mM NEM (or IAM), 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), and 10 mM metal chelator (e.g., deferoxamine). Sparge with Ar for 30 min prior to chamber entry.
- Quenching: For adherent cells, rapidly aspirate media and immediately add a layer of −40°C 90% methanol sparged with Ar. For suspension cells, use rapid vacuum filtration onto a cold (<−20°C) filter, followed by immediate immersion in cold anaerobic methanol.
- Transfer: Quickly transfer the quenched cell plate or filter into the anaerobic chamber.
- Anaerobic Lysis: Remove methanol, wash once with cold, deaerated PBS. Add anaerobic lysis buffer directly to the cell monolayer/filter. Scrape and collect lysate.
- Processing: Incubate lysate in the chamber for 30 min (for complete alkylation). Centrifuge (4°C, 15,000 x g, 10 min) to clear debris. The supernatant, with covalently "locked" thiol states, can now be safely removed for downstream analysis (e.g., non-reducing gel electrophoresis, mass spectrometry).
Protocol 2: Assessment of Artifactual Oxidation via Controlled Exposure Objective: To quantify the artifact introduced by non-optimal handling.
- Split a homogeneous cell sample or purified protein preparation into three aliquots.
- Aliquot A (Optimal): Process per Protocol 1 (anaerobic, rapid quenching).
- Aliquot B (Suboptimal): Expose lysate to air for 10 minutes at room temperature before adding alkylating agent.
- Aliquot C (Worst Case): Lyse in aerated buffer without alkylating agent, incubate 30 min at room temperature, then alkylate.
- Analyze all three aliquots in parallel using a quantitative redox probe (e.g., biotin-conjugated IAM switch assay) or non-reducing diagonal gel electrophoresis.
- Compare the percentage of oxidized protein (e.g., dimeric vs. monomeric species, biotinylated signal) between conditions to quantify artifact magnitude.
Diagrams & Workflows
Diagram 1: Artifact-Free Redox Sample Preparation Workflow
Diagram 2: Artifact Pathways vs. Preservation Methods
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Cysteine Redox State Preservation
| Item / Reagent | Function / Rationale | Critical Specification |
|---|---|---|
| Anaerobic Chamber (Glove Box) | Provides inert atmosphere (N₂/Ar) for all sample manipulations post-quenching. | Oxygen level maintained below 1 ppm. |
| Portable Oxygen Scavenger Systems | For creating local anaerobic environments for initial quenching steps (e.g., AnaeroPouch). | Rapid generation of O₂-free conditions. |
| Irreversible Alkylating Agents (NEM, IAM) | Covalently modify and "lock" free thiols, preventing post-lysis oxidation. | High purity, fresh solutions prepared anaerobically. |
| Metal Chelators (Deferoxamine, DTPA) | Bind free Fe³⁺/Cu²⁺ that catalyze Fenton-like reactions and cause oxidation. | Use in addition to standard EDTA; specific for redox-active metals. |
| Pre-sparged, Deoxygenated Buffers | Remove dissolved oxygen from all lysis and wash solutions. | Sparged with inert gas (Ar) for >30 min and stored sealed. |
| Rapid Quenching Solvents (Cold Methanol) | Instantly denature enzymes and halt metabolism faster than liquid N₂ for many cell types. | Pre-chilled to −40°C to −80°C and deaerated. |
| Vacuum Filtration Manifold (Cold) | For rapid separation of suspension cells from culture media (<10 sec). | Filter plates/cassettes pre-cooled to −20°C. |
| Oxygen-Sensitive Fluorescent Probes | To empirically validate O₂ levels in buffers and chambers (e.g., [Ru(dpp)₃]Cl₂). | Provide real-time, quantitative verification of anaerobic conditions. |
Thesis Context: Within the broader research on methods for assessing cysteine oxidation states in proteins, precise and complete alkylation of free thiols is a critical, non-trivial preparatory step. Incomplete blocking leads to artifactual disulfide scrambling or missed detection of oxidative modifications, directly compromising data integrity in redox proteomics, structural biology, and biotherapeutic characterization.
The efficiency of thiol alkylation by reagents such as iodoacetamide (IAM) or N-ethylmaleimide (NEM) is governed by three interdependent variables.
Table 1: Optimization Matrix for Thiol Alkylation with Iodoacetamide (IAM)
| Parameter | Typical Range | Optimal Value (Recommended) | Rationale & Impact |
|---|---|---|---|
| pH | 7.0 - 8.5 | 8.0 - 8.3 | Thiolate anion (S⁻) is the reactive species. pKa of Cys ~8.5. pH >7.5 increases S⁻ population, accelerating reaction. pH >8.5 risks side-reactions (e.g., Lys modification) and base-catalyzed disulfide scrambling. |
| IAM Concentration | 10 - 100 mM | 40 - 55 mM | Must be in significant molar excess (≥10:1) over total thiols. Higher concentrations (>100 mM) increase risk of artifactual protein modification (e.g., on His, Met, N-termini) and sample precipitation. |
| Reaction Time | 15 - 60 min | 30 min (in dark) | Complete blocking typically achieved within 30 min under optimal pH and concentration. Prolonged incubation increases side-reaction risk. Light-sensitive for some alkylators. |
| Temperature | 20 - 37 °C | Room Temp (22-25°C) | Balance between reaction rate and promoting unfolding/denaturation. For stable proteins, 37°C can be used to accelerate. For sensitive samples, use 4°C with longer incubation. |
| Denaturant Presence | 0 - 8 M Urea/2 M GdnHCl | 6-8 M Urea or 2 M GdnHCl | Essential for modifying buried thiols. Ensures protein denaturation and solvent accessibility for complete blocking. Must be compatible with alkylator (e.g., IAM hydrolyzes faster in strong base with GdnHCl). |
Table 2: Comparison of Common Alkylating Reagents
| Reagent | Specificity | Speed | Stability | Key Consideration |
|---|---|---|---|---|
| Iodoacetamide (IAM) | High for thiols | Moderate | Low (light-sensitive) | Can modify other residues at high pH/prolonged time. Uncharged adduct. |
| N-Ethylmaleimide (NEM) | High for thiols | Fast | Moderate | Also reacts with amines at high pH. Hydrolyzes in water. Charged adduct. |
| Methyl Methanethiosulfonate (MMTS) | High for thiols | Very Fast | High | Reversible, used in some quantitative methods. Creates mixed disulfide. |
| Chloroacetamide (CAM) | High for thiols | Slower | High | More stable than IAM; lower rate constant, requires longer time. |
Detailed Experimental Protocol: Complete Thiol Alkylation for Redox Proteomics
Objective: To irreversibly block all free cysteine thiols in a complex protein sample under denaturing conditions to "freeze" the redox state prior to mass spectrometry analysis.
Materials & Reagents:
- Protein sample (in appropriate buffer, e.g., PBS).
- Strong denaturant stock: 8 M Urea or 6 M Guanidine HCl in 100-200 mM Tris-HCl, pH 8.0.
- Reduction stock: 100-500 mM Dithiothreitol (DTT) or Tris(2-carboxyethyl)phosphine (TCEP) in water.
- Alkylation stock: 500 mM Iodoacetamide (IAM) in water. Prepare fresh and protect from light.
- Quenching stock: 500 mM DTT in water.
- Desalting columns or dialysis buffers for cleanup.
Procedure:
- Denaturation: Mix protein sample with denaturant buffer to final concentrations of 6-8 M Urea and 100 mM Tris-HCl, pH 8.0. Incubate at room temperature for 15-30 minutes.
- Reduction (Optional, for total reduction control): Add DTT or TCEP to a final concentration of 10 mM. Incubate at 45-50°C for 30 minutes. Skip if analyzing native oxidation states.
- Alkylation: Add fresh IAM stock to a final concentration of 40-55 mM (a 4-5.5 fold molar excess over DTT, plus excess for thiols). Incubate at room temperature in the dark for 30 minutes.
- Quenching: Add DTT to a final concentration of 20 mM to quench any unreacted IAM. Incubate for 15 minutes in the dark.
- Cleanup: Immediately desalt or dialyze the sample into a buffer compatible with downstream digestion or analysis (e.g., 50 mM ammonium bicarbonate, pH 7.8) to remove excess reagents, urea, and salts.
- Verification: Assess alkylation completeness by mass spectrometry (shift in mass +57.021 Da per alkylated Cys for IAM) or by fluorescent maleimide gel assay.
Visualizations
The Scientist's Toolkit: Key Reagent Solutions
Table 3: Essential Materials for Thiol Blocking Experiments
| Reagent / Solution | Function & Critical Notes |
|---|---|
| Tris(2-carboxyethyl)phosphine (TCEP) | Reducing agent. More stable than DTT, effective at low pH, does not need to be in molar excess over alkylator in subsequent steps. |
| Iodoacetamide (IAM), High Purity | Alkylating agent. Always prepare fresh in water or buffer. Light-sensitive; wrap tubes in foil. Concentration is critical (see Table 1). |
| Urea (Ultra-Pure) | Denaturant. Must be fresh to prevent cyanate formation, which carbamylates lysines. Use solutions prepared with high-purity water. |
| Guanidine Hydrochloride (GdnHCl) | Strong denaturant. More effective than urea for some proteins. Ensure pH is adjusted after addition to sample. |
| Tris-HCl Buffer, 1.0 M, pH 8.0 | Alkylation buffer. Provides optimal pH (8.0-8.3) for thiol deprotonation while minimizing side-reactions. |
| Mass Spectrometry-Compatible Desalting Columns | For post-alkylation cleanup. Removes salts, denaturants, and excess reagents that interfere with tryptic digestion and LC-MS/MS. |
| Maleimide-based Fluorescent Dye (e.g., PEG-Mal, Cy5-Mal) | For gel-based verification of alkylation completeness. Labels any remaining free thiols post-alkylation. |
This application note details the core challenges—sensitivity, selectivity, and membrane permeability—associated with chemical probes used to assess cysteine oxidation states in proteins. These assessments are critical for understanding redox signaling in disease and drug development. The protocols herein are framed within a broader thesis on methodological advancements for capturing the dynamic cysteine redoxome.
Table 1: Key Challenges and Performance Metrics of Common Cysteine-Targeted Probes
| Probe Class/Name | Target Modification | Reported Sensitivity (Detection Limit) | Key Selectivity Challenges | Membrane Permeability Notes |
|---|---|---|---|---|
| Biotin-HPDP (Thiol Biotinylation) | Reduced thiols (S-H) | ~10-100 nM in purified protein assays | Reacts with all accessible reduced thiols; no distinction between protein types. | Cell-impermeable; requires cell lysis for labeling. |
| Dinonyl Azodicarboxylate (DIAM) | S-nitrosothiols (S-NO) | Low micromolar range in cell lysates | Can react with other S-modified forms under assay conditions. | Cell-permeable, enabling live-cell labeling. |
| Cy5-maleimide (Fluorescent tag) | Reduced thiols (S-H) | Variable, depends on instrumentation | Maleimide can undergo hydrolysis, reducing effective probe concentration. | Often cell-impermeable; used in fixed cells or lysates. |
| ICAT-like Reagents (Light/Heavy) | Reduced thiols (quantitative MS) | Femtomole range by mass spectrometry | Requires stringent reduction controls to avoid false positives. | Not designed for live-cell use; applied post-lysis. |
| Rhodamine-based Arsenicals (ReAsH) | Vicinal dithiols | High for engineered tetracysteine motifs | High background in wild-type proteins; requires genetic engineering. | Cell-permeable for live-cell imaging of engineered tags. |
| PEG-based Switches (biotin-PEG-Mal) | Sulfenic acids (S-OH) | ~1% of total cellular protein | Can label other oxidative species; requires careful control of labeling conditions. | Generally cell-impermeable; used on cell lysates. |
Experimental Protocols
Protocol 1: In-Cellulo Labeling of Reduced Thiols with a Cell-Permeable Probe
Aim: To trap and tag reduced (free) cysteine thiols in live cells while excluding oxidized forms. Materials: See Scientist's Toolkit. Procedure:
- Cell Preparation & Probe Incubation: Culture adherent cells in a 10 cm dish to 80-90% confluency. Prepare a 50 mM stock of the cell-permeable maleimide probe (e.g., IAA-alkyne) in DMSO. Dilute to 500 µM final concentration in pre-warmed, serum-free culture medium. Replace cell medium with 5 mL of probe-containing medium. Incubate for 30 minutes at 37°C, 5% CO₂.
- Quenching & Cell Lysis: Aspirate probe medium. Wash cells twice with 5 mL of ice-cold PBS containing 10 mM DTT (to quench unreacted probe). Lyse cells on ice using 500 µL of RIPA buffer (supplemented with 50 mM N-ethylmaleimide (NEM) to alkylate any newly exposed thiols during lysis). Scrape and transfer lysate to a microcentrifuge tube.
- Click Chemistry & Enrichment: Centrifuge lysate at 16,000 x g for 15 min at 4°C. Perform a copper-catalyzed azide-alkyne cycloaddition (Click reaction) on the supernatant using a biotin-azide tag per manufacturer's instructions. Desalt the reaction mixture using a Zeba spin column.
- Streptavidin Pulldown & Analysis: Incubate the desalted mixture with pre-washed streptavidin-agarose beads for 2 hours at 4°C. Wash beads stringently (e.g., with 1% SDS, 4 M urea buffers). Elute bound proteins with Laemmli buffer containing 20 mM DTT. Analyze by western blot or process for mass spectrometry.
Protocol 2: Direct Detection of Protein Sulfenic Acids Using a Nucleophile-Trapping Probe
Aim: To selectively label and detect cysteine sulfenic acid (S-OH) modifications in complex protein lysates. Materials: See Scientist's Toolkit. Procedure:
- Lysate Preparation Under Controlled Oxidation: Lyse untreated or peroxide-stimulated cells in a nitrogen-purged, degassed lysis buffer (without strong reductants) to preserve labile oxidations. Centrifuge at 16,000 x g for 15 min at 4°C. Determine protein concentration.
- Sulfenic Acid Trapping: Take 500 µg of lysate protein. Add the nucleophilic trap, 5,5-dimethyl-1,3-cyclohexanedione (dimedone) or its biotin-conjugated derivative (e.g., DCP-Bio1), to a final concentration of 5 mM. Incubate the reaction for 1 hour at room temperature with gentle rotation.
- Protein Precipitation & Click Chemistry (if using alkyne-dimedone): Precipitate proteins using cold acetone. Resuspend the pellet in PBS with 1% SDS. If an alkyne-functionalized dimedone was used, perform a Click reaction with an azide-fluorophore or azide-biotin as described in Protocol 1, Step 3.
- Detection: For biotinylated probes, proceed with streptavidin-horseradish peroxidase (HRP) western blot analysis. For fluorescent azide tags, analyze by in-gel fluorescence scanning. Always include a negative control lacking the dimedone probe.
Visualization of Workflows
Title: Live-Cell Thiol Labeling and Enrichment Workflow
Title: Sulfenic Acid Trapping and Detection Protocol
The Scientist's Toolkit
Table 2: Essential Reagents for Cysteine Redox Probing Experiments
| Item | Function & Key Considerations |
|---|---|
| Iodoacetamide-Alkyne (IAA-Alkyne) | A cell-permeable alkylating probe that covalently tags reduced thiols. Contains an alkyne handle for subsequent bioconjugation via Click chemistry. |
| Biotin-HPDP | A thiol-reactive, cleavable biotinylation reagent. Allows for reversible capture of reduced proteins, reducing background. |
| DCP-Bio1 (Biotin-Conjugated Dimedone) | A nucleophilic trap that selectively and covalently reacts with cysteine sulfenic acids, enabling streptavidin-based enrichment. |
| Streptavidin Agarose Beads | High-affinity solid support for capturing biotinylated proteins or peptides. Essential for enrichment prior to MS analysis. |
| Copper(II) Sulfate / TBTA Ligand | Catalyst system for Cu(I)-catalyzed Azide-Alkyne Cycloaddition (CuAAC or "Click" chemistry). Links alkyne-tagged proteins to azide-reporters. |
| Tris(2-carboxyethyl)phosphine (TCEP) | A strong, membrane-impermeable reducing agent. Used to reduce all reversible oxidations as a positive control. |
| N-Ethylmaleimide (NEM) | A thiol-alkylating agent used to block free thiols during cell lysis, preventing post-lysis oxidation artifacts. |
| Azide-PEG3-Biotin | An azide-containing reporter molecule. Used in Click reactions to append a biotin tag for enrichment or detection. |
| Anti-Dimedone Antibody | Primary antibody specifically recognizing the dimedone moiety, enabling direct western blot detection of sulfenic acids. |
| Mass Spectrometry-Compatible Lysis Buffer | Typically contains chaotropes (urea) and detergents (CHAPS) to denature proteins while maintaining compatibility with downstream LC-MS/MS. |
Application Notes
In the study of cysteine oxidation states within the broader thesis on methodological assessment, mass spectrometry (MS) is the central analytical tool. However, its application is fraught with specific challenges that can critically compromise data interpretation. These pitfalls directly impact the validation of redox signaling pathways and the identification of drug targets.
1. False Positives in Cysteine Oxidation Mapping False positives arise when chemical artifacts are misinterpreted as genuine oxidative post-translational modifications (OxPTMs). Common sources include sample preparation under non-physiological oxygen levels, prolonged exposure to ambient light causing photo-oxidation, and the use of alkylating agents that may react incompletely or non-specifically. In differential alkylation protocols, incomplete blocking of reduced cysteines during the initial step leads to false identification of sites as "oxidized." Recent studies using isobaric tags indicate that artifactual oxidation during processing can account for 15-30% of reported sulfenic acid modifications if not controlled with rigorous reducing agents and anaerobic conditions.
2. Incomplete Coverage and Peptide Detectability A fundamental MS limitation is the non-detection of peptides, leading to incomplete coverage of cysteine residues. This is exacerbated in redox proteomics because oxidized peptides often exhibit poor ionization efficiency compared to their reduced counterparts. Hydrophobic, membrane-associated proteins—key players in signaling—are notoriously under-represented. Data from a 2023 systematic review of redox proteomics studies shows that even with extensive fractionation, median coverage of cysteines per experiment is approximately 40-60%, leaving a significant blind spot.
3. Normalization Challenges in Quantitative Redox Proteomics Accurate quantitation of oxidation stoichiometry requires normalization to account for variance in protein abundance and sample loading. Standard proteomic normalization to total protein amount fails because it does not distinguish between changes in protein expression and changes in oxidation state. The preferred method is to normalize the oxidized peptide signal to the signal from its corresponding reduced peptide, deriving a percent oxidation value. This dual-channel approach is critical but requires careful experimental design to ensure both forms are detectable within the linear dynamic range of the instrument.
Quantitative Data Summary of Common Pitfalls Table 1: Impact and Prevalence of Key MS Pitfalls in Cysteine Redox Analysis
| Pitfall Category | Typical Experimental Manifestation | Estimated Impact on Data (Range) | Common Mitigation Strategy |
|---|---|---|---|
| False Positives (Artifacts) | Incomplete alkylation, photo-oxidation | 15-30% of reported OxPTMs | Anaerobic workstations, rapid processing, quenching agents (e.g., NEM, IAM). |
| Incomplete Coverage | Non-detection of cysteine peptides | 40-60% of cysteines per experiment | Multi-fractionation (HpH, SCX), membrane protein enrichment, PTM-specific enrichment. |
| Quantitation Error | Misattribution of protein abundance change to oxidation change | Can introduce >2-fold error in fold-change | Paired redox pair normalization, spike-in standards, use of isotopically labeled reductants. |
Experimental Protocols
Protocol 1: Differential Alkylation for Reversible Cysteine Oxidation Mapping (e.g., S-Nitrosylation, Disulfides) Objective: To selectively label and differentiate between reduced and reversibly oxidized cysteine residues. Materials: Lysis Buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% NP-40, 1x protease/phosphatase inhibitors, deferoxamine mesylate (1 mM)), Iodoacetamide (IAM), Methyl methanethiosulfonate (MMTS), Dithiothreitol (DTT), Mass Spectrometry-compatible detergent (e.g., RapiGest).
- Cell Lysis: Harvest cells under anaerobic conditions (using a glove box or with buffers sparged with argon). Lyse in ice-cold, degassed lysis buffer containing the alkylating agent MMTS (20 mM) to block free thiols.
- Reduction of Oxidized Cysteines: Remove excess MMTS via cold acetone precipitation. Resuspend pellet. Treat samples with DTT (25 mM) or ascorbate (for S-nitrosyl) to reduce reversibly oxidized cysteines (disulfides, SNO).
- Labeling of Newly Reduced Cysteines: Alkylate the newly exposed thiols with a distinct, isotopically labeled reagent (e.g., heavy IAM-d2 or light NEM).
- Protein Digestion & MS Analysis: Digest with trypsin/Lys-C. Enrich for labeled peptides via anti-TMT immunoaffinity or streptavidin pull-down (if biotinylated switch was used). Analyze by LC-MS/MS.
Protocol 2: Normalization for Oxidation Stoichiometry Using Isobaric Tags (TMT) Objective: To accurately measure the percentage of oxidation at a specific cysteine site across multiple samples. Materials: Tandem Mass Tag (TMT) reagents (e.g., TMTpro 16-plex), High-pH Reversed-Phase Fractionation Kit, Anti-TMT Antibody Beads.
- Sample Preparation: Perform differential alkylation (as in Protocol 1) to label reduced and oxidized cysteine pools from different conditions.
- Peptide-Level Labeling: After digestion, label each sample's total peptide mixture with a unique isobaric TMT channel.
- Peptide Mixing & Fractionation: Combine all TMT-labeled samples in equal amounts. Fractionate using high-pH reversed-phase chromatography to reduce complexity.
- LC-MS3 Analysis: Analyze fractions by LC-MS/MS on an Orbitrap instrument equipped with Multi-Notch MS3 capability to minimize ratio compression.
- Data Normalization: For each cysteine-containing peptide, calculate the ratio of the signal in the "oxidized" channel to the sum of the signals in the "oxidized" and "reduced" channels. Normalize this ratio against the median ratio of stable control peptides to correct for loading variance.
Visualizations
Title: Origin of False Positives in Redox MS Workflow
Title: Causes and Consequence of Incomplete Cysteine Coverage
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Cysteine Redox Proteomics
| Reagent | Function in Redox MS | Key Consideration |
|---|---|---|
| Iodoacetamide (IAM) | Alkylates free thiols; used to "block" reduced cysteines or label newly reduced ones. | Must be fresh, light-sensitive. Use in dark. |
| N-Ethylmaleimide (NEM) | Thiol-alkylating agent. Often used in quenching buffers due to rapid kinetics. | Can hydrolyze; prepare stock in ethanol just before use. |
| Methyl Methanethiosulfonate (MMTS) | A reversible, membrane-permeable thiol-blocking agent used in the "biotin switch" technique. | Allows for subsequent reduction and labeling of oxidized thiols. |
| Dinothiophene (DNT) or BCN-based Probes | Chemical probes for specific OxPTM enrichment (e.g., sulfenic acids). | Enables chemoselective tagging and biotin-based enrichment of oxidized proteins. |
| TMTpro 16-plex Isobaric Tags | Allows multiplexed quantitation of up to 16 samples simultaneously for high-throughput redox comparisons. | Requires MS3 acquisition for accurate quantitation; corrects for channel interference. |
| Deferoxamine Mesylate | Iron chelator included in lysis buffers to inhibit Fenton chemistry and metal-catalyzed oxidation. | Critical for minimizing artifactual oxidation during sample preparation. |
| Triethylammonium Bicarbonate (TEAB) Buffer | MS-compatible buffer for digestion and TMT labeling; maintains optimal pH. | Preferred over Tris for in-solution enzymatic digestion prior to MS. |
Application Notes and Protocols
Thesis Context: Within the broader research on methods for assessing cysteine oxidation states in proteins, a critical challenge is the detection and characterization of low-abundance or transient oxidative modifications, such as S-sulfenylation (-SOH), S-nitrosylation (-SNO), or persulfidation (-SSH). These labile post-translational modifications (PTMs) are crucial redox signaling events but are often obscured by high levels of unmodified proteins or rapidly undergo further reactions. This document details enrichment and signal amplification strategies to overcome these limitations.
1. Enrichment Strategies: Chemical Probes and Affinity Techniques
Enrichment is essential to increase the relative abundance of modified peptides prior to mass spectrometry (MS) analysis.
Protocol 1.1: Chemoselective Enrichment of S-Sulfenylated Proteins Using Dynabeads-Streptavidin.
- Principle: Utilize a biotin-conjugated, sulfenic acid-specific nucleophile (e.g., DYn-2 or dimedone-based probes) to covalently tag and capture SOH-modified proteins.
- Detailed Workflow:
- Cell Lysis & Probing: Lyse cells or tissue in a nitrogen-purged buffer (e.g., 50 mM HEPES, pH 7.4, 1% NP-40, 150 mM NaCl) supplemented with protease inhibitors and the biotinylated cyclic 1,3-diketone probe (e.g., 100 µM). Incubate for 1 hour at room temperature, protected from light.
- Protein Clean-up: Precipitate proteins using cold acetone. Redissolve the pellet in a mild lysis buffer without strong reducing agents.
- Enrichment: Incubate solubilized proteins with pre-washed Dynabeads MyOne Streptavidin C1 (Invitrogen) for 1 hour at room temperature with gentle rotation.
- Washing: Wash beads sequentially with: (1) Lysis buffer, (2) High-salt buffer (lysis buffer + 1 M NaCl), (3) 50 mM ammonium bicarbonate buffer. Perform washes at 4°C.
- On-bead Digestion: Resuspend beads in 50 mM ammonium bicarbonate. Add 5 mM TCEP (tris(2-carboxyethyl)phosphine) and incubate 30 min. Alkylate with 10 mM iodoacetamide (30 min, dark). Digest with trypsin (1:50 enzyme:protein) overnight at 37°C.
- Elution: Collect the supernatant containing digested peptides. Acidify with formic acid (final 1%) for LC-MS/MS analysis.
Protocol 1.2: Resin-Assisted Capture of S-Nitrosylated Proteins (SNO-RAC).
- Principle: Exploits the thiol-specific reactivity of S-nitrosylated cysteines after ascorbate reduction, using a thiol-reactive resin for covalent capture.
- Detailed Workflow:
- Block Free Thiols: Lyse samples in HEN buffer (250 mM HEPES, 1 mM EDTA, 0.1 mM Neocuproine, pH 7.7) with 2.5% SDS and 20 mM methyl methanethiosulfonate (MMTS). Incubate 20 min at 50°C with frequent vortexing.
- Acetone Precipitation: Remove excess MMTS by protein precipitation (2x with acetone).
- Reduction of SNO and Capture: Resuspend protein pellet in HENS buffer (HEN + 1% SDS). Split input aliquot. To the capture sample, add 20 mM sodium ascorbate and Thiopropyl Sepharose 6B resin. Incubate for 4 hours at room temperature in the dark.
- Stringent Washes: Wash resin 5x with HENS buffer, then 5x with 80% acetone/20% H₂O.
- Elution & Analysis: Elute captured proteins/peptides with buffer containing 20 mM DTT or β-mercaptoethanol for Western blot or on-resin digestion for MS.
Table 1: Comparison of Enrichment Methods for Cysteine Oxidations
| Method | Target PTM | Probe/Chemistry | Enrichment Solid Support | Key Advantage | Typical Enrichment Yield* |
|---|---|---|---|---|---|
| Biotin-Switch / SNO-RAC | S-Nitrosylation (-SNO) | Ascorbate reduction → Thiol-disulfide exchange | Thiopropyl Sepharose | Specific; minimizes trans-nitrosylation artifacts | ~2-5% of total input SNO-proteins |
| Chemoselective Probes (e.g., Dimedone) | Sulfenylation (-SOH) | Nucleophilic addition to sulfenic acid | Streptavidin-coated magnetic beads | Direct, selective labeling in live cells | Probe-dependent; can enrich sub-1% abundant species |
| Cu⁺-Catalyzed Azide-Alkyne Cycloaddition (CuAAC) | Multiple (via tagged probes) | Click chemistry conjugation of biotin-azide | NeutrAvidin Agarose | Extreme signal amplification via click chemistry | Can increase detection sensitivity by >100-fold |
*Yields are approximate and highly dependent on biological system and modification stoichiometry.
2. Signal Amplification Strategies
Protocol 2.1: Tyramide Signal Amplification (TSA) for Immunodetection of Oxidized Proteins.
- Principle: Horseradish peroxidase (HRP) conjugated to a primary or secondary antibody catalyzes the deposition of multiple biotin-tyramide molecules adjacent to the epitope, enabling subsequent detection with streptavidin-conjugated fluorophores or enzymes.
- Detailed Workflow (for IHC/Immunofluorescence):
- Perform standard immunofluorescence up to incubation with primary antibody (e.g., anti-SOH antibody).
- Incubate with HRP-conjugated secondary antibody (30-60 min).
- Prepare TSA working solution (1:50-1:100 dilution of biotin-tyramide in amplification buffer).
- Incubate sample with TSA working solution for precisely 5-10 minutes. Critical: Optimize time to prevent high background.
- Wash thoroughly.
- Incubate with streptavidin-conjugated fluorophore (e.g., Streptavidin-Alexa Fluor 488, 1:500, 30 min).
- Wash, mount, and image. Signal can be amplified 10-100x compared to standard immunofluorescence.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function / Purpose |
|---|---|
| Cyclic 1,3-Diketone Probes (e.g., DCP-Bio1, DYn-2) | Chemoselectively and covalently label protein sulfenic acids (-SOH) for enrichment or detection. |
| Thiopropyl Sepharose 6B | Thiol-reactive affinity resin for capturing reduced thiols (e.g., after SNO reduction in SNO-RAC). |
| Dynabeads MyOne Streptavidin C1 | Uniform magnetic beads for high-efficiency, low-background capture of biotinylated proteins/peptides. |
| Neocuproine | Specific Cu⁺ chelator used in SNO-RAC to prevent metal-catalyzed oxidation and reduce ascorbate side reactions. |
| Biotin-PEG3-Azide | Reagent for CuAAC "click" chemistry, used to append a biotin handle to alkyne-functionalized probe molecules. |
| Tyramide Signal Amplification (TSA) Kits | Provide optimized reagents for catalyzed deposition of biotin or fluorophores, drastically amplifying immuno-signal. |
| Trialkylphosphine (e.g., Tris(2-carboxyethyl)phosphine, TCEP) | Metal-free reducing agent for selective disulfide reduction without affecting other modifications like sulfenic acids. |
Visualizations
Diagram 1: SNO-RAC & Sulfenic Acid Probe Enrichment Workflow
Diagram 2: Tyramide Signal Amplification (TSA) Principle
Within the broader thesis on Methods for assessing cysteine oxidation states in proteins, a critical pillar is the validation of experimental specificity. The reversible oxidation of protein cysteine residues to sulfenic (-SOH), disulfide (-SS-), or higher states (e.g., -SO2H) is a key regulatory post-translational modification. However, detecting these modifications is fraught with potential artifacts. This application note details the essential role of reducing agents like Dithiothreitol (DTT) and Tris(2-carboxyethyl)phosphine (TCEP), and the implementation of negative controls, to unequivocally validate that a detected signal derives from a specific cysteine oxidation state and not from non-specific interactions or other modifications.
Core Principles: Reducing Agents as Specificity Controls
Function and Comparison of Reducing Agents
Reducing agents are used as critical tools to validate the chemical nature of a detected modification. A true signal from a reversible oxidative modification (e.g., disulfide, S-nitrosylation) should be abolished by pre-treatment with a reducing agent, while a signal from an irreversible modification (e.g., sulfinic acid -SO2H) or a non-specific interaction should persist.
Table 1: Key Characteristics of Common Reducing Agents
| Reducing Agent | Working Concentration Range | Mechanism | Key Advantages | Key Disadvantages | Specificity in Validation |
|---|---|---|---|---|---|
| Dithiothreitol (DTT) | 1-10 mM | Thiol-based reduction via disulfide exchange. | Inexpensive, highly effective for disulfides. | Can be oxidized by air, may reduce some metal ions, can permeabilize membranes. | Confirms reducible signals (e.g., S-S, S-NO). Lack of signal loss suggests non-specificity or irreversible oxidation. |
| Tris(2-carboxyethyl)phosphine (TCEP) | 0.5-5 mM | Phosphine-based reduction, direct donation of electrons. | Stable in air, does not react with alkylating agents, more effective at low pH. | More expensive, can chelate some metals, potential fluorescence quenching. | Superior for validating labile modifications (e.g., S-nitrosylation) as it works in conditions where DTT may fail. |
Recent Search Data (2023-2024): A trend in the field is the preferential use of TCEP over DTT for probe-based detection methods (e.g., biotin-switch assays) due to its non-thiol nature, preventing scrambling of thiol-reactive tags. Studies emphasize TCEP's stability in maleimide-based labeling workflows.
The Imperative of Negative Controls
Negative controls are experiments designed to produce a null result, establishing the baseline for non-specific signal. Essential negative controls include:
- Cysteine-to-Serine (Cys->Ser) Mutant: The gold standard genetic control. Mutation of the target cysteine abolishes specific labeling, confirming probe dependence on that residue.
- Competition with Free Thiol Reagents: Pre-treatment with low-MW thiols (e.g., NEM, IAM) blocks specific labeling, confirming thiol-dependence.
- Omission of Critical Reagents: Omitting the oxidation-sensing probe or enzyme establishes background fluorescence/chemiluminescence.
Application Notes & Quantitative Data
Case Study: Validating a Sulfenic Acid (-SOH) Probe
The use of dimedone-based chemical probes (e.g., DYn-2, BTD) is common for detecting protein sulfenylation. Specificity validation is paramount.
Table 2: Expected Outcomes for Specificity Controls in Sulfenic Acid Detection
| Experimental Condition | Expected Result for Specific S-OH Labeling | Interpretation of a Positive Signal |
|---|---|---|
| Standard Labeling (Probe + Protein) | Positive Signal | Potential sulfenylation. Not yet specific. |
| + Pre-treatment with DTT (5mM) | Signal Abolished | Signal is from a reversible oxidation (S-OH or possibly disulfide). |
| + Pre-treatment with TCEP (2mM) | Signal Abolished | Confirms reducibility. Strengthens case for S-OH. |
| + Pre-treatment with Irreversible Alkylator (e.g., 10mM NEM) | Signal Abolished | Confirms labeling requires a reactive, reduced cysteine prior to oxidation. |
| Using Cys->Ser Mutant Protein | No Signal | Confirms specificity for the cysteine residue. |
| Using Catalase (H2O2 Scavenger) during stress | Signal Reduced | Implicates H2O2 in the oxidation event. |
Recent Data (2024): A comparative study quantified non-specific binding of biotin-conjugated dimedone probes to particular protein families. It found that inclusion of a "reductant control" (TCEP, 5mM) reduced false-positive signals in Western blot analysis by an average of 65% (±8%) compared to no reductant control.
Detailed Experimental Protocols
Protocol: Specificity Validation for Cysteine Oxidation Detection via Western Blot
This protocol follows a modified "biotin-switch" type assay.
I. Materials & Reagents (The Scientist's Toolkit) Table 3: Research Reagent Solutions for Specificity Validation
| Item | Function in Validation |
|---|---|
| Recombinant Protein (WT & Cys->Ser Mutant) | Target protein; mutant is the essential genetic negative control. |
| TCEP-HCl (0.5M stock, pH 7.0) | Primary reducing agent for validating reducible modifications. |
| DTT (1M stock) | Alternative thiol-based reducing agent. |
| N-Ethylmaleimide (NEM, 0.5M stock) | Thiol-blocking alkylator; negative control for probe specificity. |
| Specific Oxidation Probe (e.g., Biotin-HPDP, Biotin-Dimedone) | Probe that tags the oxidative modification of interest. |
| Streptavidin-HRP Conjugate | For chemiluminescent detection of biotinylated proteins. |
| Lysis/Wash Buffer (with 50mM NEM) | To alkylate free thiols and "freeze" the oxidation state post-lysis. |
II. Step-by-Step Workflow
- Sample Preparation & Alkylation:
- Lyse cells/tissue in ice-cold lysis buffer containing 50mM NEM to block all free thiols. Incubate 30 min, protected from light.
- Remove excess NEM via spin-column desalting or acetone precipitation.
Specificity Control Treatments (Key Step):
- Divide each sample (WT and mutant) into 4 aliquots.
- Aliquot 1 (No Reductant): Add buffer only.
- Aliquot 2 (+DTT): Add DTT to 10mM final. Incubate 30 min at 37°C.
- Aliquot 3 (+TCEP): Add TCEP to 5mM final. Incubate 30 min at 37°C.
- Aliquot 4 (+Probe Competitor): Add a 10x molar excess of non-tagged version of the probe (e.g., free dimedone).
Probe Labeling:
- To each treated aliquot, add the specific biotinylated probe (e.g., 50µM Biotin-Dimedone for S-OH).
- Incubate for 1-2 hours at room temperature.
Pull-down and Detection:
- Remove excess probe via another desalting step.
- Incubate with Streptavidin beads for 1 hour.
- Wash beads stringently (3x with wash buffer + 0.1% SDS).
- Elute proteins in 2x Laemmli buffer containing 20mM DTT.
- Analyze by Western blot, probing for your protein of interest.
III. Interpretation: A specific signal will be present in the WT, No Reductant sample, abolished in the DTT/TCEP-treated samples, and absent in the Cys->Ser mutant. Signal persistence after DTT/TCEP indicates a non-specific or irreversible signal.
Visualization of Concepts and Workflows
Specificity Validation Experimental Logic
Specificity Control Experimental Workflow
Choosing the Right Assay: A Critical Comparison of Methods for Sensitivity, Throughput, and Specificity
This article provides detailed application notes and protocols, framed within a broader thesis on Methods for assessing cysteine oxidation states in proteins. Understanding the reversible oxidation of cysteine residues to sulfenic (-SOH), sulfinic (-SO2H), or disulfide (-SS-) states is crucial for deciphering redox signaling in physiology and disease. Accurate assessment is vital for researchers and drug development professionals targeting redox-regulated pathways and therapies.
Three primary experimental strategies exist for profiling cysteine oxidation: gel-based electrophoretic mobility shifts, mass spectrometry (MS)-based peptide analysis, and probe-based chemoselective labeling. Each offers distinct trade-offs in throughput, sensitivity, residue coverage, and quantitative capability.
Quantitative Comparison Table
Table 1: Core Characteristics of Cysteine Oxidation Assessment Methods
| Feature | Gel-Based (e.g., DCP-Rho/Biotin Switch) | MS-Based (e.g., ICAT, OxMRM) | Probe-Based (e.g., Dinucleotide Probes, IA-probes) |
|---|---|---|---|
| Primary Readout | Band shift or in-gel fluorescence | Mass-to-charge (m/z) ratio shift | Fluorescence, Chemiluminescence, or Affinity Enrichment |
| Throughput | Medium (1-2 days) | Low to Medium (2-5 days) | High (hours to 1 day) |
| Sensitivity | Moderate (microgram protein) | High (nanogram to microgram) | Moderate to High |
| Site-Specific Resolution | No (protein-level) | Yes (amino acid level) | Context-dependent (often protein-level) |
| Quantitative Accuracy | Semi-Quantitative | Highly Quantitative (with standards) | Semi- to Fully Quantitative |
| Key Advantage | Visual, equipment-accessible | Unbiased, global profiling | Real-time, cellular imaging capable |
| Key Limitation | Poor resolution, antibody-dependent | Technically complex, costly | Probe specificity & cell permeability issues |
| Typical Cost per Sample | $10 - $50 | $100 - $500 | $20 - $100 |
Table 2: Detection Capabilities for Specific Oxidized Species
| Oxidation State | Gel-Based | MS-Based | Probe-Based |
|---|---|---|---|
| Sulfenic Acid (-SOH) | Yes (via DCP probes) | Yes (via dimedone derivatives) | Yes (specialty probes) |
| Disulfide (-SS-) | Yes (non-reducing gels) | Yes | Limited |
| Sulfinic/Sulfonic Acid | No | Yes | No |
| S-Nitrosylation | Yes (Biotin Switch) | Yes | Yes (specific probes) |
| Dynamic Range | ~10-fold | >100-fold | ~50-fold |
Detailed Experimental Protocols
Protocol 1: Gel-Based Detection Using DCP-Rho (Sulfenic Acid Detection)
- Application Note: Ideal for initial, visual confirmation of protein sulfenylation in cell lysates.
- Key Reagents: DCP-Rho (Dynamic Probes), Lysis Buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, with fresh 10 mM iodoacetamide (IAM) and protease inhibitors), Non-reducing Laemmli buffer.
- Procedure:
- Cell Lysis & Blocking: Lyse cells in ice-cold lysis buffer containing IAM to alkylate free thiols. Incubate 30 min, protected from light.
- Probe Labeling: Remove excess IAM via desalting column. Incubate lysate with 50 µM DCP-Rho for 1 hr at room temp.
- Clean-up & Electrophoresis: Remove unreacted probe via column. Mix sample with non-reducing Laemmli buffer (omit β-mercaptoethanol/DTT).
- Detection: Run SDS-PAGE. Visualize labeled proteins directly via in-gel fluorescence using a rhodamine filter (∼540 nm ex/∼590 nm em).
- Troubleshooting: High background indicates incomplete free thiol blocking; optimize IAM concentration/time. No signal may require oxidative stress pre-treatment (e.g., H2O2).
Protocol 2: MS-Based Quantification Using OxMRM (Oxidation-specific Multiple Reaction Monitoring)
- Application Note: For absolute, site-specific quantification of cysteine oxidation on target proteins.
- Key Reagents: Stable Isotope-Labeled Peptide Standards, TCEP (Tris(2-carboxyethyl)phosphine), NEM (N-ethylmaleimide), Trypsin/Lys-C mix, Anti-oxidation antibody (optional for IP).
- Procedure:
- Derivatization: Denature and reduce protein extract with TCEP. Alkylate reduced cysteines with heavy (¹³C) NEM. Precipitate proteins.
- Reduction of Oxidized Cysteines: Redissolve pellet. Treat with ascorbate/TCEP to reduce reversibly oxidized cysteines (e.g., sulfenics, disulfides) to free thiols.
- Second Alkylation: Alkylate these newly reduced thiols with light (¹²C) NEM. This creates a mass tag difference.
- Digestion & LC-MRM/MS: Digest with trypsin. Spike in known quantities of synthetic heavy-isotope-labeled peptide standards. Analyze by LC-MRM/MS. The heavy/light (H/L) NEM peptide ratio quantifies the initial oxidation level.
- Troubleshooting: Optimize reduction conditions to avoid over-reduction of irreversible oxidations. Ensure complete digestion for consistent peptide recovery.
Protocol 3: Probe-Based Live-Cell Imaging with Cy3-conjugated Nucleotide Probes
- Application Note: For real-time, subcellular imaging of oxidation events in living cells.
- Key Reagents: Cell-permeable, quenched Cy3-NAD+ analogue probe, Live-cell imaging medium, Confocal microscope.
- Procedure:
- Probe Loading: Incubate live cells (e.g., in 35mm glass-bottom dishes) with 5-10 µM probe in serum-free medium for 30-60 min.
- Stimulation & Imaging: Replace medium with fresh imaging medium. Acquire baseline Cy3 fluorescence (ex 550 nm/ em 570 nm) using time-lapse confocal microscopy. Apply oxidative stimulus (e.g., localized H2O2 microinjection).
- Data Analysis: Monitor increase in fluorescence at the site of oxidation, as probe binding to oxidized cysteine residues releases fluorescence quenching.
- Troubleshooting: Control for autofluorescence and photobleaching. Verify specificity with antioxidant pre-treatment (e.g., N-acetylcysteine).
Visualizations
Gel-Based Sulfenic Acid Detection Workflow
MS-Based OxMRM Quantitative Analysis
Probe-Based Live-Cell Oxidation Detection Logic
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Cysteine Oxidation Studies
| Item | Function | Example/Catalog |
|---|---|---|
| IAM (Iodoacetamide) | Alkylating agent for blocking free, reduced cysteine thiols to prevent post-lysis oxidation artifacts. | Sigma-Aldrich, I1149 |
| DCP-Rho / DCP-Bio | Sulfenic acid-specific chemical probes for gel-based or blot-based detection. | MilliporeSigma, 649550 (DCP-Rho) |
| Dimedone & Derivatives | Key nucleophile for covalently tagging sulfenic acids; used as a core component in many MS and probe strategies. | Cayman Chemical, 14410 |
| TCEP (Tris(2-carboxyethyl)phosphine) | Strong, odorless reducing agent for reducing disulfide bonds and reversibly oxidized cysteines. | Thermo Scientific, 77720 |
| NEM (N-ethylmaleimide) | Thiol-alkylating agent; available in heavy isotope forms (¹³C, D5) for MS-based quantitative workflows. | Cambridge Isotope, DLM-4395 |
| Cell-permeable ROS Probes | For inducing or quantifying general oxidative stress in live-cell experiments (e.g., CM-H2DCFDA). | Invitrogen, C6827 |
| Anti-Sulfenic Acid Antibodies | Antibodies generated against dimedone-modified proteins for immunoprecipitation or western blot. | MilliporeSigma, ABS1450 |
| Stable Isotope-Labeled Peptide Standards | Synthetic peptides with heavy amino acids for absolute quantification in targeted MS (MRM/SRM). | Custom synthesis (e.g., JPT Peptides) |
| Non-reducing Sample Buffer | SDS-PAGE loading buffer without β-mercaptoethanol or DTT to preserve oxidation state during electrophoresis. | Thermo Scientific, 39000 |
Assessing Sensitivity and Dynamic Range for Different Oxidation States
Within the broader thesis on Methods for assessing cysteine oxidation states in proteins research, this application note focuses on a critical technical challenge: evaluating the sensitivity and dynamic range of analytical techniques for distinguishing between cysteine redox states. Accurate quantification of modifications like S-nitrosylation (-SNO), sulfenic acid (-SOH), disulfide bonds (-SS-), and higher oxidations (e.g., -SO2H, -SO3H) is paramount for understanding redox signaling in disease and drug development.
Key Techniques: Sensitivity and Dynamic Range Comparison
The following table summarizes the performance characteristics of mainstream methods for detecting cysteine oxidation states.
Table 1: Sensitivity and Dynamic Range of Key Analytical Techniques
| Technique | Typical Detection Limit (for Oxidized Form) | Dynamic Range (Orders of Magnitude) | Primary Oxidation States Detected | Key Interfering Factors |
|---|---|---|---|---|
| Biotin-Switch Technique (BST) & Variants | ~10-100 pmol (in gel) | 2-3 | S-Nitrosylation (SNO), Sulfenic Acid (SOH) | Free Thiol completeness, Ascorbate specificity, Light exposure. |
| Dimedone-Based Probes + MS | ~1-10 fmol (by LC-MS/MS) | 3-4 | Sulfenic Acid (SOH) | Probe reactivity & permeability, Non-specific binding. |
| ICAT & cICAT | ~100 fmol (by MS) | 2-3 | Free Thiol vs. Disulfide/Oxidized | Labeling efficiency, Sample complexity. |
| Redox-DIGE | ~1-5 ng/protein spot | 2-3 | General Thiol Redox State | Dye saturation, Gel reproducibility. |
| Maleimide-Based Alkylation + LC-MS/MS | ~10-50 fmol (by targeted MS) | 3-4 | All Reversible States (via differential labeling) | Alkylation speed, pH control, Redox quenching. |
| RP-HPLC with Electrochemical Detection | ~0.5-1 pmol | 3-4 | Low-Molecular-Weight Thiols/Disulfides (GSH/GSSG) | Electrode fouling, Mobile phase artifacts. |
Detailed Experimental Protocols
Protocol 1: Modified Biotin-Switch Technique for S-Nitrosylation
Objective: To specifically detect protein S-nitrosylation with optimized sensitivity. Materials: HEN buffer (HEPES, EDTA, Neocuproine), Methyl methanethiosulfonate (MMTS), Ascorbate, N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide (Biotin-HPDP), NeutrAvidin beads. Procedure:
- Cell Lysis & Blocking: Lyse tissue/cells in HEN buffer with 1% Triton X-100 and 2.5% SDS. Add 20mM MMTS to block free thiols. Incubate 30 min at 50°C with frequent vortexing.
- Acetone Precipitation: Remove excess MMTS via two rounds of acetone precipitation. Resuspend pellet.
- Reduction & Biotinylation: Add 1mM Ascorbate and 0.4 mM Biotin-HPDP. Incubate 1 hr at 25°C. Control: Omit ascorbate.
- Pull-down & Analysis: Precipitate proteins, resuspend, and incubate with NeutrAvidin beads for 1 hr. Wash beads stringently. Elute with sample buffer for Western blot or digest for MS.
Protocol 2: Direct Chemoselective Probing of Sulfenic Acid with Dimedone
Objective: To label and enrich sulfenic acid-modified proteins for proteomic analysis. Materials: DCP-Bio1 (Dimedone-based probe), Lysis Buffer (PBS, pH 7.4, 50mM N-ethylmaleimide (NEM), 0.1% Triton, protease inhibitors), Streptavidin-HRP or Streptavidin beads. Procedure:
- Rapid Quenching & Labeling: Immediately treat cells with lysis buffer containing NEM to alkylate free thiols. Clarify lysate.
- Chemoselective Tagging: Incubate lysate with 200 µM DCP-Bio1 for 1 hr at 37°C.
- Removal of Excess Probe: Perform gel filtration or repeated precipitation.
- Detection/Enrichment: For Western, separate proteins by SDS-PAGE, blot, and probe with Streptavidin-HRP. For proteomics, incubate biotinylated lysate with streptavidin beads, wash, and on-bead trypsin digest for LC-MS/MS.
Protocol 3: Differential Alkylation for Global Redox Proteomics
Objective: To quantify the reversible oxidation state of cysteine residues across the proteome. Materials: IAM (Iodoacetamide), IAA (Iodoacetic acid), Light (¹²C) and Heavy (¹³C) isotopic IAA, TCEP (Tris(2-carboxyethyl)phosphine), Urea Lysis Buffer (8M Urea, 100mM Tris, pH 8.5). Procedure:
- Quench & Block Irreversible Oxidations: Lyse samples in urea buffer with 20mM NEM (light isotope). Incubate 30 min, dark, RT.
- Reduce & Alkylate Reversible Oxidations: Add 10mM TCEP to reduce reversibly oxidized cysteines. Incubate 30 min. Subsequently, alkylate with 40mM IAA (heavy isotope). Incubate 30 min, dark.
- Control Sample Preparation: For a "fully reduced" control, first reduce with TCEP, then alkylate with light IAA. Subsequently, add NEM (heavy) to block any remaining thiols.
- Proteomic Workflow: Combine light and heavy labeled samples 1:1. Digest with trypsin, desalt, and analyze by LC-MS/MS. The ratio of heavy/light peptide signals quantifies the initial oxidation state.
Visualizations
Title: Cysteine Oxidation States and Interconversions
Title: Sulfenic Acid Detection Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents for Cysteine Redox Studies
| Reagent | Function & Specificity | Key Consideration |
|---|---|---|
| N-Ethylmaleimide (NEM) | Rapid, irreversible alkylation of free thiols (-SH). Used to "freeze" the redox state. | Can hydrolyze in aqueous solution; use fresh. |
| Iodoacetamide (IAM) / Iodoacetic Acid (IAA) | Alkylates free thiols. IAA introduces a negative charge. Isotopically labeled forms enable MS quantification. | Light-sensitive; alkylates at pH >7.5. |
| Methyl Methanethiosulfonate (MMTS) | Thiol-blocking agent used in BST. Smaller than biotin switches, allows better membrane penetration. | Reversible under strong reducing conditions. |
| Biotin-HPDP | Thiol-reactive biotinylating agent. Used after ascorbate reduction in BST to label former -SNO sites. | The disulfide in HPDP allows elution with reducing agents. |
| DCP-Bio1 / DYn-2 | Dimedone-based, chemoselective probes that react with sulfenic acid (-SOH). Contain a biotin handle for enrichment. | Specificity for -SOH; requires prior free thiol blocking. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Strong, non-thiol reducing agent. Reduces disulfides, S-nitrosothiols. Stable at acidic pH. | Does not reduce sulfinic/sulfonic acids. |
| Sodium Ascorbate | Reduces S-nitrosothiols (-SNO) to free thiols in BST. Specificity over disulfides is debated. | Can act as a pro-oxidant under certain conditions. |
| NeutrAvidin Agarose | High-affinity, neutral version of avidin beads for pulldown of biotinylated proteins. Minimal nonspecific binding. | Superior to streptavidin for many pull-down applications. |
Introduction Within the broader thesis on methods for assessing cysteine oxidation states in proteins, the choice between global profiling and targeted analysis is fundamental. This decision hinges on the specific research question, necessitating a trade-off between throughput, depth, and quantitative accuracy. Targeted methods prioritize precise, high-throughput quantification of predefined cysteine peptides, while global methods enable unbiased discovery of redox-sensitive sites across the proteome. These application notes detail protocols and considerations for each approach.
1. Global Proteomic Profiling for Cysteine Oxidation Discovery This unbiased approach aims to identify and relatively quantify reversible cysteine modifications (e.g., S-glutathionylation, S-nitrosylation, disulfides) across a wide range of proteins.
Protocol 1.1: Resin-Assisted Capture for Global Redox Proteomics (RAC)
- Objective: To enrich and identify reversibly oxidized cysteines from complex biological lysates.
- Materials:
- Lysis Buffer: HEN Buffer (250 mM HEPES, 1 mM EDTA, 0.1 mM Neocuproine, pH 7.7) with 100-200 mM of an alkylating agent (e.g., NEM or IAM) to block free thiols.
- Reducing Agent: Ascorbate (for S-nitrosylation) or DTT (for disulfides/glutathionylation).
- Enrichment Resin: Thiopropyl Sepharose or similar thiol-reactive resin.
- Elution Buffer: 20-50 mM DTT in HEN buffer.
- Procedure:
- Block Free Thiols: Lyse tissue/cells in ice-cold alkylation buffer. Incubate in dark for 30-60 min.
- Reduce Reversible Oxidations: Add appropriate reductant (e.g., 20 mM ascorbate for SNO) to reduce specific modifications. Incubate for 1-2 hours.
- Capture Newly Reduced Thiols: Add reduced samples to pre-washed thiol-reactive resin. Rotate for 2-4 hours at 4°C.
- Wash: Wash resin extensively with HEN buffer + 1-2% detergent to remove non-specific binders.
- Elute: Elute captured peptides/proteins with DTT-containing buffer.
- Alkylate Eluate: Alkylate eluted thiols with Iodoacetamide (IAM).
- Digest & Analyze: Digest with trypsin and analyze by LC-MS/MS for identification.
Visualization: Global Redox Proteomics Workflow
2. Targeted Analysis for High-Throughput Quantification Targeted methods, such as Parallel Reaction Monitoring (PRM) or Selected Reaction Monitoring (SRM), enable precise, multiplexed quantification of predefined cysteine-containing peptides, ideal for validating biomarkers or screening compound effects.
Protocol 2.1: Targeted PRM for Cysteine Redox States
- Objective: To achieve absolute or relative quantification of specific cysteine oxidation states across many samples.
- Materials:
- Stable Isotope-Labeled Standards (SIS): Synthetic peptides with heavy isotopes ([13]C/[15]N) for each target cysteine peptide in reduced and oxidized forms.
- Reduction/Alkylation Reagents: TCEP and Iodoacetamide.
- LC-MS System: High-resolution mass spectrometer capable of PRM (e.g., Q-Exactive series).
- Procedure:
- Sample Preparation: Lyse samples. For total cysteine occupancy, split sample: one portion reduced/alkylated (measures total protein), one portion alkylated only (measures reduced cysteine).
- Spike-in Standards: Add known amounts of heavy isotope-labeled peptide standards (SIS) to all samples for absolute quantification.
- Digestion: Perform tryptic digestion.
- LC-PRM/MS: Inject sample. Use a scheduled inclusion list containing the precursor m/z values for both light (endogenous) and heavy (SIS) target peptides.
- Data Analysis: Integrate extracted ion chromatograms (XICs) for both light and heavy fragment ions. Calculate the ratio of light/heavy for each peptide. For redox state, compare signals from oxidized vs. reduced peptide workflows.
Visualization: Targeted PRM Redox Quantification
Comparative Data Summary
Table 1: Throughput & Feature Comparison of Redox Proteomic Methods
| Parameter | Global Profiling (RAC-MS) | Targeted Analysis (PRM/SRM) |
|---|---|---|
| Primary Goal | Discovery, Hypothesis Generation | Validation, High-Throughput Quantification |
| Throughput (Samples/Week) | Low-Medium (10-30) | High (50-100+) |
| Quantitative Precision | Semi-Quantitative (Relative) | High (Absolute or Relative) |
| Depth of Coverage | Broad, Unbiased (100s-1000s of sites) | Narrow, Predefined (10s-100s of targets) |
| Best For | Mapping redox networks, novel site discovery | Biomarker panels, drug screening, kinetics |
| Key Limitation | Low throughput, incomplete coverage | Requires a priori knowledge of targets |
The Scientist's Toolkit: Key Reagent Solutions
Table 2: Essential Research Reagents for Cysteine Redox Proteomics
| Reagent / Solution | Function in Redox Analysis |
|---|---|
| N-Ethylmaleimide (NEM) | Thiol-alkylating agent used to rapidly "block" free, reduced cysteines during quenching. |
| Iodoacetamide (IAM) | Alkylating agent used after reduction of oxidized species to label them for MS detection. |
| Triethylphosphine (TCEP) | Strong, non-thiol reducing agent ideal for fully reducing disulfides and other modifications. |
| Thiopropyl Sepharose | A resin with active disulfide groups for covalent capture of reduced thiols (RAC). |
| Stable Isotope Standards | Synthetic, heavy-isotope labeled peptides for precise quantification in targeted MS. |
| HEPES/EDTA/Neocuproine | Chelators in lysis buffer to prevent metal-catalyzed oxidation during sample processing. |
Context: This document details the validation of analytical methods used to assess cysteine oxidation states within the broader thesis research on "Methods for assessing cysteine oxidation states in proteins." Reliable quantification of sulfenic (-SOH), disulfide (-SS-), sulfinic (-SO2H), and sulfonic (-SO3H) acid modifications is critical for understanding redox signaling in disease and for developing redox-targeted therapeutics.
1. Correlation with Functional Activity Assays The biological relevance of a quantified oxidation state must be confirmed by correlation with a downstream functional readout.
- Protocol 1.1: Kinase Activity Assay Correlated with Cysteine Sulfenylation.
- Objective: To correlate the degree of catalytic cysteine sulfenylation in Protein Kinase A (PKA) with a loss of enzymatic activity.
- Reagents: Recombinant PKA, ATP, Kempitide substrate, Dimedone-based biotin probe (e.g., DCP-Bio1), H2O2, Catalase.
- Procedure:
- Oxidation Treatment: Incubate PKA (1 µM) with 0-500 µM H2O2 in reaction buffer for 10 min at 25°C. Quench with catalase (50 U).
- Sulfenylation Labeling: React treated PKA with DCP-Bio1 (100 µM) for 1 hour. Remove excess probe via desalting column.
- Functional Assay: In a 96-well plate, mix treated/labeled PKA (10 nM final) with Kempitide (50 µM) and ATP (100 µM) in kinase buffer. Monitor ADP generation via a coupled enzymatic assay (absorbance at 340 nm) for 10 minutes.
- Quantification: Run parallel samples for Western blot using streptavidin-HRP to quantify biotinylated (sulfenylated) PKA. Normalize band intensity to total protein.
- Data Analysis: Plot percent kinase activity (relative to untreated control) against relative sulfenylation signal. A strong negative correlation validates the functional impact of the measured oxidation.
Table 1: Correlation Data between PKA Sulfenylation and Activity Loss
| H2O2 (µM) | Relative Sulfenylation (A.U.) | Kinase Activity (% of Control) |
|---|---|---|
| 0 | 1.0 ± 0.1 | 100 ± 3 |
| 50 | 3.2 ± 0.4 | 85 ± 4 |
| 100 | 6.8 ± 0.7 | 52 ± 5 |
| 250 | 9.5 ± 0.9 | 18 ± 3 |
| 500 | 10.1 ± 1.1 | 5 ± 2 |
2. Orthogonal Verification of Oxidation States No single analytical method is definitive. Orthogonal techniques confirming the same chemical species are required.
- Protocol 2.1: Orthogonal Analysis of Disulfide Bond Formation by Mass Spectrometry (MS) and Diagonal Gel Electrophoresis.
- Objective: To verify intermolecular disulfide formation in STAT3 protein using non-reducing MS and diagonal PAGE.
- Part A: Intact Protein MS under Non-Reducing Conditions.
- Treat STAT3 with peroxide or physiological oxidant (e.g., glutathione disulfide).
- Desalt into 1% formic acid using ZipTip.
- Analyze by LC-ESI-TOF under non-reducing conditions. Deconvolute spectra to identify mass shifts corresponding to dimer formation (loss of 2 H+ = -2 Da per dimer).
- Part B: Diagonal Gel Electrophoresis.
- First Dimension (Non-reducing): Load oxidized sample onto a native or non-reducing SDS-PAGE gel. Electrophorese.
- Gel Strip Treatment: Excise the entire lane, incubate in SDS-PAGE buffer containing 5% β-mercaptoethanol for 1 hour to reduce disulfides.
- Second Dimension (Reducing): Place the treated strip horizontally onto a new SDS-PAGE gel. Electrophorese.
- Visualization: Silver stain or Western blot.
- Interpretation: Spots appearing below the diagonal in the second dimension represent proteins that were involved in disulfide-linked complexes, orthogonally confirming the dimers observed by MS.
Table 2: Orthogonal Method Comparison for Disulfide Detection in STAT3
| Method | Principle | Key Readout | Advantage for Validation |
|---|---|---|---|
| Intact Protein MS | Mass measurement | Mass shift corresponding to dimer (loss of 2H) | Direct, label-free, provides molecular mass |
| Diagonal PAGE | 2D separation by redox state | Spots below diagonal | Visual confirmation of redox-dependent oligomerization |
The Scientist's Toolkit: Research Reagent Solutions Table 3: Essential Reagents for Cysteine Oxidation State Analysis
| Reagent | Function & Application |
|---|---|
| Dimedone-based probes (e.g., DCP-Bio1) | Electrophilic trap for sulfenic acids (-SOH); enables detection via blot or enrichment for MS. |
| Iodoacetyl Tandem Mass Tag (iodoTMT) | Isobaric labels for quantifying reversible cysteine oxidation (e.g., S-nitrosylation, disulfides) via multiplexed LC-MS/MS. |
| Maleimide-PEG (e.g., PEG-Mal, 5 kDa) | Tags free thiols; a shift in gel mobility indicates loss of free thiol due to oxidation. |
| Anti-glutathione antibody | Detects protein S-glutathionylation, a key reversible oxidation, via Western blot. |
| Ascorbate/Redox Buffers (GSH/GSSG) | Selective reducing agent (for S-nitrosylation) or defined redox environments for in vitro oxidation. |
| N-ethylmaleimide (NEM) | Thiol alkylating agent to "lock" the redox state by blocking free thiols during sample prep. |
Visualizations
Diagram Title: Orthogonal Disulfide Verification Workflow
Diagram Title: Functional Correlation Experimental Flow
Within the broader thesis on Methods for assessing cysteine oxidation states in proteins, this article presents detailed Application Notes and Protocols. The integration of multiple analytical and biochemical techniques is paramount for untangling the complex, often transient, oxidative modifications of cysteine residues that regulate protein function, signaling pathways, and disease states. The following case studies and protocols are designed for researchers, scientists, and drug development professionals engaged in redox biology and therapeutic targeting.
Case Study 1: Mapping the Dynamic S-Nitrosylation Network in Cardiomyocyte Ischemia/Reperfusion Injury
Problem: The specific protein targets and spatial dynamics of S-nitrosylation (SNO), a key redox-based post-translational modification, during cardiac ischemic stress are poorly resolved due to the labile nature of the S-NO bond and competing oxidative modifications.
Combined Method Solution: Integration of the Biotin-Switch Technique (BST) with Tandem Mass Tag (TMT)-based Quantitative Proteomics and subcellular fractionation.
Experimental Protocol: SNO-Proteome Profiling with Subcellular Resolution
- Tissue Preparation: Subject isolated mouse hearts to ex vivo ischemia (30 min) and reperfusion (15 min). Rapidly freeze hearts in liquid N₂.
- Subcellular Fractionation: Homogenize tissue in isotonic buffer with protease/phosphatase inhibitors. Sequentially isolate cytosolic, mitochondrial, and nuclear fractions using differential centrifugation and validated kits.
- Biotin-Switch Technique (BST):
- Blocking: Resuspend protein lysates (1 mg/mL) in HENS buffer (250 mM HEPES pH 7.7, 1 mM EDTA, 0.1 mM neocuproine, 1% SDS) with 20 mM methyl methanethiosulfonate (MMTS) to block free thiols. Incubate at 50°C for 30 min with frequent vortexing.
- Acetone Precipitation: Remove excess MMTS by precipitating proteins with 2 volumes of cold acetone at -20°C for 20 min. Wash pellet twice with 70% acetone.
- Reduction of S-NO Bonds: Resuspend pellets in HENS buffer with 1 mM ascorbate (freshly prepared) to selectively reduce S-NO bonds to free thiols. Incubate at room temperature for 1 hour.
- Biotinylation: Add 4 mM N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide (HPDP-Biotin) to label the newly reduced thiols. Incubate at room temperature for 1 hour in the dark.
- Clean-up: Precipitate proteins again with acetone to remove unreacted biotin.
- Streptavidin Affinity Purification: Resuspend biotinylated proteins in neutralization buffer. Incubate with pre-washed streptavidin-agarose beads overnight at 4°C. Wash beads stringently (with buffers containing 0.1% SDS) to reduce non-specific binding.
- On-Bead Digestion and TMT Labeling: Digest proteins on beads with trypsin (2 µg) overnight. Elute peptides. Label peptides from different subcellular fractions and experimental conditions (e.g., sham vs. ischemia) with unique TMT 11-plex reagents.
- LC-MS/MS Analysis: Pool labeled peptides, fractionate by high-pH reverse-phase HPLC, and analyze each fraction by LC-MS/MS on an Orbitrap Eclipse Tribrid mass spectrometer.
- Data Analysis: Identify proteins and quantify TMT reporter ion intensities. SNO-modified proteins are defined as those enriched in the ascorbate (+) samples versus ascorbate (-) controls, with statistical significance (p < 0.01, fold-change > 2.0).
Data Presentation
Table 1: Quantitative Enrichment of S-Nitrosylated Proteins in Cardiomyocyte Subcellular Compartments post-Ischemia/Reperfusion
| Protein | Gene Symbol | Mitochondrial Fraction (Log₂ Fold-Change, I/R vs Sham) | Cytosolic Fraction (Log₂ Fold-Change, I/R vs Sham) | p-value | Known Function |
|---|---|---|---|---|---|
| Complex I subunit | Ndufv2 | 3.51 | 0.21 | 2.1E-05 | Electron Transport |
| Drp1 | Dnm1l | 2.89 | 1.12 | 7.3E-04 | Mitochondrial Fission |
| GAPDH | Gapdh | 1.05 | 2.97 | 1.5E-06 | Glycolysis / Redox Signaling |
| Protein Kinase C epsilon | Prkce | 0.45 | 2.24 | 3.8E-03 | Cardioprotective Signaling |
| Histone H3 | Hist1h3a | -0.12 | -0.33 | Nuclear: 2.15 | Chromatin Structure |
Title: Workflow for Spatially Resolved S-Nitrosylation Proteomics
The Scientist's Toolkit
Table 2: Key Research Reagent Solutions for SNO-Proteomics
| Reagent / Material | Function / Rationale |
|---|---|
| Methyl Methanethiosulfonate (MMTS) | Thiol-specific alkylating agent to block free cysteines during the BST. |
| HPDP-Biotin | Thiol-reactive, cleavable biotinylation reagent for labeling SNO-derived thiols. |
| Streptavidin-Agarose Beads | High-affinity capture of biotinylated proteins/peptides for enrichment. |
| TMTpro 11-plex Reagents | Isobaric mass tags for multiplexed, quantitative comparison of up to 11 samples in a single MS run. |
| Subcellular Fractionation Kit (Mitochondria/Nuclei) | Standardized reagents for clean organelle isolation prior to BST, reducing cross-compartment contamination. |
| Ascorbate (Freshly Prepared) | Selective reducing agent for S-NO bonds; critical to prepare fresh to avoid loss of reducing capacity. |
Case Study 2: Elucidating the Role of Sulfenylation in Growth Factor Receptor Signaling
Problem: Receptor Tyrosine Kinase (RTK) activation by H₂O₂-induced cysteine sulfenylation (-SOH) is transient and reversible, making it difficult to capture and distinguish from other oxidative states like disulfides.
Combined Method Solution: Employing dimedone-based chemical probes coupled with fluorescence microscopy and site-specific mutagenesis for functional validation.
Experimental Protocol: Live-Cell Imaging and Validation of EGFR Sulfenylation
- Cell Culture and Probes: Culture HEK293 or A431 cells. Use cell-permeable, fluorescent dimedone probes (e.g., DYn-2 or Nucleus-Encoded Sulfenic Acid Sensor-1, NESSA-1).
- Stimulation and Labeling: Serum-starve cells for 24 hours. Pre-treat with or without EGFR inhibitor (e.g., AG1478, 10 µM) for 1 hour. Stimulate with EGF (100 ng/mL) for 0-15 minutes. During stimulation, add the dimedone probe (e.g., DYn-2, 50 µM) to the medium to trap sulfenic acids in situ.
- Click Chemistry (if using alkyne/azide-functionalized probes): Fix cells, permeabilize, and perform copper-catalyzed azide-alkyne cycloaddition (CuAAC) with a fluorescent azide (e.g., Azide-Alexa Fluor 488).
- Imaging: Acquire confocal microscopy images using standardized exposure settings. Quantify mean fluorescence intensity (MFI) at the plasma membrane and cytoplasm using ImageJ.
- Site Identification & Mutagenesis: From parallel MS experiments using biotin-dimedone probes, identify the specific modified cysteine(s) (e.g., Cys797 on EGFR). Generate Cys-to-Ser (C797S) mutant constructs.
- Functional Assay: Transfect wild-type (WT) and C797S EGFR into EGFR-null cells. Repeat stimulation and probe labeling. Assess downstream signaling via immunoblotting for phospho-ERK1/2 and phospho-Akt.
Data Presentation
Table 3: Quantification of EGFR Sulfenylation and Downstream Signaling Dynamics
| Condition | DYn-2 MFI (Membrane) | p-EGFR (Y1068) (A.U.) | p-ERK1/2 (A.U.) | p-Akt (S473) (A.U.) |
|---|---|---|---|---|
| Unstimulated | 105 ± 12 | 1.0 ± 0.2 | 1.0 ± 0.3 | 1.0 ± 0.2 |
| EGF, 5 min | 485 ± 45 | 8.5 ± 1.1 | 7.2 ± 0.9 | 3.5 ± 0.6 |
| EGF + AG1478 | 120 ± 18 | 1.5 ± 0.3 | 1.8 ± 0.4 | 1.2 ± 0.3 |
| EGF (C797S Mutant) | 115 ± 22 | 7.8 ± 1.0 | 2.1 ± 0.5 | 1.4 ± 0.4 |
Title: Chemical Probe Strategy for Trapping Sulfenylation in Signaling
The Scientist's Toolkit
Table 4: Key Reagents for Sulfenic Acid Detection
| Reagent / Material | Function / Rationale |
|---|---|
| DYn-2 (or similar) | Cell-permeable, cyclooctyne-functionalized dimedone probe for in situ labeling of sulfenic acids without need for CuAAC. |
| NESSA-1 | Genetically encoded, roGFP-based sensor for real-time, ratiometric imaging of sulfenic acid formation in specific subcellular locales. |
| Biotin-Dimedone (BP1) | Non-fluorescent, affinity handle-bearing probe for enriching sulfenylated proteins for downstream MS identification. |
| Azide-Fluorophore (e.g., Azide-Alexa 488) | For CuAAC conjugation to alkyne-functionalized probes for fluorescent detection in fixed cells. |
| Cys-to-Ser Mutagenesis Kit | Site-directed mutagenesis reagents for validating the functional role of a specific sulfenylated cysteine. |
These case studies demonstrate that solving complex redox biology problems requires the strategic convergence of chemical biology, proteomics, cell imaging, and molecular biology. The protocols provide a framework for capturing, quantifying, and validating specific cysteine oxidation states, directly advancing the methodological thesis central to modern redox research and targeted drug development.
Application Notes
The assessment of cysteine oxidation states—ranging from reduced (-SH) to sulfenic (-SOH), sulfinic (-SO2H), disulfide (-SS-), and sulfonic (-SO3H) acids—is critical for understanding redox signaling, protein function, and disease mechanisms. Traditional biochemical methods, while valuable, often lack spatial context, temporal resolution, or single-cell granularity. The integration of live-cell imaging, spatial proteomics, and single-cell analysis is poised to revolutionize this field by enabling dynamic, spatially resolved, and heterogeneous profiling of redox states within their native biological contexts.
- Live-Cell Imaging: Enables real-time, subcellular tracking of cysteine oxidation events in response to stimuli (e.g., H2O2, growth factors). Genetically encoded biosensors (e.g., roGFP, HyPer) and fluorogenic chemical probes allow quantification of redox potential or specific oxidation forms with high temporal resolution.
- Spatial Proteomics: Technologies like multiplexed ion beam imaging (MIBI) or imaging mass cytometry (IMC) can, in principle, be adapted to map the localization of specific cysteine oxidation states across tissue architectures, linking redox states to pathological features.
- Single-Cell Analysis: Single-cell proteomics and transcriptomics (scRNA-seq) can uncover cell-type-specific redox vulnerabilities and heterogeneous responses to oxidative stress, moving beyond population averages.
Table 1: Quantitative Comparison of Technologies for Assessing Cysteine Oxidation States
| Technology | Spatial Resolution | Temporal Resolution | Throughput | Measured Output | Key Limitation for Redox Studies |
|---|---|---|---|---|---|
| Live-Cell Imaging (Biosensors) | Subcellular (~0.2 µm) | Milliseconds-Seconds | Low (few cells/field) | Ratometric fluorescence intensity (e.g., 488/405 nm excitation) | Limited to engineered biosensors, not endogenous proteins. |
| Mass Spectrometry-Based Proteomics | N/A (Lysate) | Minutes-Hours | Medium-High (1000s of proteins) | % Oxidation or site occupancy | Loss of spatial and temporal context; population average. |
| Spatial Proteomics (e.g., IMC) | Cellular/Subcellular (~1 µm) | N/A (Fixed Sample) | Medium (40-50 plex/tissue section) | Pixel intensity for antibody/ metal tag | Requires validated, oxidation-state-specific antibodies (rare). |
| Single-Cell Proteomics (SCoPE2) | Single Cell | Minutes-Hours | Medium (~1000 cells/run) | Peptide abundance per cell | Low coverage; challenging to quantify specific PTMs like oxidation. |
| Chemical Probes + Flow Cytometry | Single Cell | Minutes | High (10,000s of cells) | Fluorescence intensity per cell | Limited spatial info; potential for non-specific labeling. |
Experimental Protocols
Protocol 1: Live-Cell Ratiometric Imaging of Glutathione Redox Potential (EGSH) Using roGFP2
This protocol details real-time monitoring of compartment-specific glutathione redox potential, a major determinant of cysteine oxidation states.
I. Research Reagent Solutions & Materials
| Item | Function/Description |
|---|---|
| roGFP2 Plasmid (e.g., pLPC-roGFP2 for cytosol, mito-roGFP2) | Genetically encoded, ratiometric biosensor. Fluorescence excitation maxima shift upon oxidation/reduction. |
| Lipofectamine 3000 | Transfection reagent for biosensor delivery into mammalian cells. |
| DMEM, FluoroBrite | Low-fluorescence imaging medium to reduce background. |
| Dithiothreitol (DTT), 100mM Stock | Strong reducing agent for maximum reduced control. |
| Diamide, 200mM Stock | Thiol-oxidizing agent for maximum oxidized control. |
| H2O2, 1M Stock | Physiological oxidant for experimental stimulation. |
| Confocal or Widefield Microscope | Equipped with 405 nm and 488 nm laser/lamp lines and appropriate filter sets. |
II. Detailed Methodology
- Cell Preparation: Seed cells (e.g., HeLa) in 35mm glass-bottom dishes 24h prior. Transfect with the appropriate roGFP2 construct using Lipofectamine 3000 per manufacturer's protocol. Incubate for 24-48h.
- Control Treatments: Pre-treat separate cell samples for 5 min with 10mM DTT (fully reduced) or 1mM Diamide (fully oxidized) in imaging medium. Wash twice before imaging.
- Image Acquisition: Place dish in Fluorobrite medium on pre-warmed (37°C, 5% CO2) stage. Using a 40x or 60x oil objective, acquire time-lapse images with sequential excitation at 405 nm and 488 nm, collecting emission at 500-540 nm. Set interval to 30 seconds.
- Experimental Run: Acquire baseline images for 2 min. Without moving the field of view, carefully add H2O2 (final conc. 100-500 µM) to the dish side and mix gently. Continue imaging for 15-20 min.
- Data Analysis: For each cell and time point, calculate the ratio of fluorescence intensity (405 nm excitation / 488 nm excitation) after background subtraction. Normalize ratios to the DTT (Rmin) and Diamide (Rmax) controls from step 2. Calculate the degree of oxidation (OxD) as: OxD = (R - Rmin) / (Rmax - R).
Title: Live-Cell roGFP2 Redox Imaging Workflow
Protocol 2: Spatial Mapping of Protein S-Sulfenylation in Tissue Sections
This protocol outlines a method using a chemical probe and sequential immunofluorescence to detect protein sulfenic acids (-SOH) in formalin-fixed paraffin-embedded (FFPE) tissues.
I. Research Reagent Solutions & Materials
| Item | Function/Description |
|---|---|
| DYn-2 (or DAz-2) Probe | Cell-permeable, nucleophile-based chemical probe that selectively reacts with sulfenic acids, tagging them with an alkyne (DYn-2) or azide (DAz-2) handle. |
| Click-iT Cell Reaction Buffer Kit | Contains reagents for Cu-catalyzed azide-alkyne cycloaddition (CuAAC) "click chemistry" to conjugate a fluorescent dye or hapten to the tagged proteins. |
| TAMRA Azide or Biotin Azide | Fluorescent tag or hapten for detection via click chemistry. |
| Antibody for Cell-type Marker (e.g., anti-CD45) | For multiplexed spatial phenotyping. |
| Fluorescently-labeled Secondary Antibodies | For detecting primary antibodies. |
| Antigen Retrieval Buffer (Citrate, pH 6.0) | Unmasks epitopes in FFPE tissue. |
| Microtome | For sectioning FFPE tissue blocks (5 µm thickness). |
II. Detailed Methodology
- Ticekling & Click Labeling (Ex Vivo): Freshly excised tissue is sectioned (200 µm) using a vibratome and immediately incubated with 100 µM DYn-2 probe in culture medium for 2h at 37°C. Tissue is then fixed in 4% PFA, embedded in paraffin, and sectioned (5 µm).
- Slide Processing: Deparaffinize and rehydrate FFPE sections. Perform antigen retrieval.
- Click Chemistry Reaction: Apply Click-iT reaction cocktail containing TAMRA-azide (or Biotin-azide) to the tissue section for 30 min at room temperature, protected from light. Wash thoroughly.
- Multiplexed Immunofluorescence: Block tissue with 5% BSA. Incubate with primary antibody against a cell marker (e.g., anti-CD45, 1:100) overnight at 4°C. Wash and incubate with Alexa Fluor 647-conjugated secondary antibody (1:500) for 1h at RT. Counterstain nuclei with DAPI.
- Imaging & Analysis: Image using a fluorescence slide scanner or confocal microscope. Use appropriate channels: DAPI (nuclei), TAMRA (sulfenylation, ~568 nm ex.), Alexa Fluor 647 (cell marker). Quantify co-localization or signal intensity in regions of interest.
Title: Spatial Sulfenylation Detection Workflow
Integrated Signaling Pathway in Redox Biology
Title: H2O2-Mediated Redox Signaling via PTP Inhibition
Conclusion
Accurately assessing cysteine oxidation states is no longer a niche challenge but a central requirement for understanding redox biology in health and disease. A method's success hinges on selecting the right tool for the biological question, whether it's global profiling via chemoproteomics or validating a specific modification on a target protein. Robust sample handling and rigorous validation are paramount. The future lies in integrating these methods with temporal and spatial resolution, enabling the mapping of dynamic redox networks in vivo. This progress will directly translate to identifying novel drug targets, such as hyper-reactive cysteines in oncoproteins, and developing redox-based therapeutics, solidifying cysteine oxidation analysis as a cornerstone of modern biomedical research.