Cysteine Redox Switches: From Molecular Mechanisms to Therapeutic Targeting in Human Disease
This article provides a comprehensive synthesis of the redox regulation of protein cysteine residues, a dynamic post-translational mechanism central to cellular signaling and homeostasis.
Cysteine Redox Switches: From Molecular Mechanisms to Therapeutic Targeting in Human Disease
Abstract
This article provides a comprehensive synthesis of the redox regulation of protein cysteine residues, a dynamic post-translational mechanism central to cellular signaling and homeostasis. We explore the foundational chemistry of reactive cysteines and the spectrum of their oxidative modifications, such as S-sulfenylation and S-glutathionylation. The content details cutting-edge methodological advances in redox proteomics and computational modeling for profiling the cysteine redoxome. Furthermore, it addresses key challenges in troubleshooting pathway specificity and discusses the validation of redox-sensitive proteins as promising targets for therapeutic intervention in conditions ranging from liver disease to aging and cancer, offering a roadmap for researchers and drug development professionals in the biomedical sciences.
The Chemistry and Biology of Cysteine Redox Switches
Within the intricate landscape of cellular signaling, the unique chemistry of the cysteine thiol group enables proteins to sense and transduce changes in cellular redox status. Cysteine is one of the least abundant amino acids, yet it is frequently found as a highly conserved residue within functional sites in proteins, exhibiting extreme patterns of conservation indicative of strong selective pressure [1]. This conservation stems from the singular properties of the sulfur atom in its thiol (-SH) functional group, which can undergo a remarkable array of reversible chemical modifications in response to reactive oxygen, nitrogen, and sulfur species (ROS/RNS/RSS) [2] [3]. These modifications allow specific cysteine residues to act as "redox switches," controlling critical biological processes including gene expression, enzyme activity, and cellular adaptation to stress [2] [4].
The redox-sensing capability of cysteine is not merely a chemical curiosity but a fundamental mechanism of physiological regulation. Cells continually monitor their environment and adapt to changing surroundings by coordinated changes in gene expression and metabolic pathways, with cysteine thiols serving as key environmental sensors [2]. This whitepaper examines the chemical basis for cysteine's unique reactivity, the biological mechanisms of thiol-based redox switching, experimental approaches for studying these processes, and the emerging therapeutic implications for targeting redox-sensitive pathways in human disease.
Fundamental Chemical Properties of Cysteine Thiols
The Electronic Basis of Sulfur Reactivity
The exceptional reactivity of cysteine in redox sensing originates from the electronic configuration of sulfur. Unlike methionine, which contains a less reactive thioether, cysteine features a ionizable thiol group that can undergo deprotonation to form a negatively charged thiolate anion (RS-) under physiological conditions [1]. This thiolate form is significantly more nucleophilic and reactive than its protonated counterpart, enabling attacks on electrophilic centers in oxidizing molecules.
The sulfur atom in cysteine is highly polarizable, meaning its electron cloud can be easily distorted in response to environmental influences. This polarizability enhances the ability of cysteine thiols to form transition states during redox reactions, stabilizing reaction intermediates and lowering activation energies [1] [5]. Furthermore, the sulfur atom can access multiple oxidation states, from the reduced thiol (-2) to various oxidized forms including disulfides (-1), sulfenic acids (0), sulfinic acids (+2), and sulfonic acids (+4), providing a rich chemical repertoire for biological signaling [2] [1].
Environmental Tuning of Cysteine Reactivity
The protein microenvironment profoundly influences cysteine reactivity through several mechanisms:
pKa Modulation: The protonation state of a cysteine thiol is governed by its acid dissociation constant (pKa). While typical thiol pKa values range from 8-9, protein environments can significantly lower this value through stabilization of the thiolate anion [1]. Strategic positioning near basic residues (histidine, arginine, lysine) or within helix dipoles creates electrostatic environments that favor deprotonation, increasing the concentration of the reactive thiolate species at physiological pH [1]. For example, in peroxiredoxins, hydrogen-bonding networks lower the pKa of the active site cysteine and simultaneously activate incoming peroxide substrates, resulting in reaction rates with hydrogen peroxide as high as 10ⷠ– 10⸠Mâ»Â¹ sâ»Â¹ [1].
Solvent Accessibility and Polarity: Despite classification in some hydrophobicity scales as a hydrophobic residue, cysteine exhibits polarity similar to serine [1]. Bioinformatics analyses reveal that cysteine residues show extreme conservation patterns, with high conservation in functional sites but poor conservation in solvent-exposed surfaces, reflecting evolutionary pressure to minimize unpaired surface cysteines that might undergo non-specific oxidation [1]. This selective pressure results in cysteine being both one of the most conserved and most degenerate amino acids, depending on its functional context.
Table 1: Factors Influencing Cysteine Reactivity in Proteins
| Factor | Effect on Reactivity | Example |
|---|---|---|
| Lowered pKa | Increases nucleophilicity by favoring thiolate formation | Active site cysteines in peroxiredoxins and phosphatases |
| Proximity to Basic Residues | Stabilizes thiolate anion through electrostatic interactions | Helix dipoles, histidine positioning |
| Hydrogen Bonding Networks | Activates both cysteine and substrate for reaction | Peroxiredoxin catalytic sites |
| Metal Coordination | Can dramatically alter redox potential | Iron-sulfur clusters, zinc fingers |
| Local Dielectric Constant | Influences proton transfer and charge stabilization | Hydrophobic active sites |
Cysteine Clustering: Bioinformatics studies reveal that cysteine residues frequently occur in clusters, particularly in proteins from organisms living in harsh environments [1]. This clustering facilitates disulfide bond formation and metal binding, creating specialized redox-active sites with enhanced sensitivity to oxidative changes. The spatial arrangement of multiple cysteine residues enables cooperative interactions that can fine-tune redox potential and create threshold responses to oxidative stimuli.
Biological Mechanisms of Thiol-Based Redox Sensing
The Spectrum of Cysteine Oxidative Modifications
Cysteine thiols undergo a diverse array of oxidative modifications that serve as molecular switches for redox signaling:
Disulfide Bond Formation: The formation of intra- or intermolecular disulfide bonds between cysteine residues represents a fundamental regulatory switch. This reversible modification can induce significant conformational changes that alter protein function, subcellular localization, or interaction partners [2] [4]. In bacteria, disulfide bond formation directly regulates transcription factors including OxyR, OhrR, and Spx, activating antioxidant gene expression in response to peroxide stress [2].
Sulfenic Acid Formation: The initial oxidation product of cysteine with hydrogen peroxide is cysteine sulfenic acid (-SOH). Although often transient, sulfenic acids can be stabilized in specific protein environments and serve both as sensory intermediates and regulatory modifications [2] [6]. For example, sulfenylation of Cys797 in the epidermal growth factor receptor (EGFR) enhances its tyrosine kinase activity, linking redox signaling to growth factor pathways [6].
S-Glutathionylation and S-Thiolation: Sulfenic acids can react with low molecular weight thiols such as glutathione to form mixed disulfides, a process known as S-glutathionylation [2]. This reversible modification can protect cysteine residues from irreversible overoxidation while simultaneously altering protein function. Similar S-thiolation reactions occur with other low molecular weight thiols, including mycothiol in Actinomycetes and bacillithiol in Firmicutes [2] [1].
S-Nitrosylation: Reactive nitrogen species can modify cysteine thiols to form S-nitrosothiols (R-SNO), providing a mechanism for nitric oxide-based signaling [2]. This modification occurs through multiple mechanisms, including reaction with nitric oxide (NO) or transnitrosation from low molecular weight S-nitrosothiols like GSNO.
Sulfenamide Formation: A more recently appreciated modification involves cyclization of sulfenic acid with a neighboring backbone amide nitrogen to form a cyclic sulfenamide [2]. This modification was first detected in structural studies of protein tyrosine phosphatases and has since been observed in bacterial redox sensors like Bacillus subtilis OhrR [2].
Table 2: Major Reversible Oxidative Modifications of Cysteine Thiols
| Modification | Chemical Structure | Representative Function | Reversibility |
|---|---|---|---|
| Disulfide | R-S-S-R' | Conformational switching in OxyR, Yap1 | Thioredoxin/Glutaredoxin |
| Sulfenic Acid | R-SOH | Redox sensing in EGFR, PTPs | Reduction or further oxidation |
| S-Glutathionylation | R-S-SG | Protection from overoxidation | Glutaredoxin |
| S-Nitrosylation | R-S-NO | NO signaling | Thioredoxin/Denitrosylases |
| Sulfenamide | Cyclic R-S-N | Stabilization of oxidized form | Reduction |
Structural Consequences of Thiol Oxidation
The biological impact of cysteine oxidation stems from the structural and functional changes induced in the modified protein. These changes can include:
Conformational Rearrangements: Disulfide bond formation can introduce covalent crosslinks that stabilize specific protein conformations. In the bacterial transcription factor OxyR, disulfide formation between Cys199 and Cys208 triggers a dramatic conformational change that activates transcription of antioxidant genes [2]. Similarly, oxidation of the yeast Yap1p transcription factor promotes structural changes that regulate its nuclear localization and DNA-binding activity [2].
Altered Protein-Protein Interactions: Redox modifications can create or disrupt interaction surfaces. For example, oxidation of the Nrf2-binding site on Keap1 disrupts the Nrf2-Keap1 complex, allowing Nrf2 translocation to the nucleus and activation of antioxidant response element (ARE)-mediated gene expression [2].
Modulated Catalytic Activity: Many enzymes contain redox-sensitive cysteine residues in their active sites. Protein tyrosine phosphatases (PTPs) feature a nucleophilic cysteine that exists as a thiolate anion at neutral pH; oxidation to sulfenic acid inactivates these enzymes, thereby potentiating tyrosine phosphorylation signaling cascades [4] [6].
The following diagram illustrates the major oxidative pathways of cysteine thiols and their biological consequences in redox signaling:
Experimental Approaches for Studying Redox-Sensitive Cysteines
Proteomic Methods for Redox Profiling
Advanced proteomic techniques have enabled comprehensive mapping of cysteine redox states under physiological and pathological conditions. These methods generally rely on differentially labeling reduced and oxidized cysteines followed by quantification using mass spectrometry [7]. The iodoacetyl Tandem Mass Tag (iodoacetyl-TMT) approach allows multiplexed analysis of cysteine redox states across multiple samples, enabling comparisons between different experimental conditions, genetic backgrounds, or developmental stages [7].
A typical redox proteomics workflow involves:
- Cell lysis under controlled conditions to preserve native redox states
- Blocking free thiols with alkylating agents
- Reducing oxidized cysteines (disulfides, sulfenic acids)
- Labeling newly reduced thiols with isotopically distinct tags
- Protein digestion, peptide purification, and mass spectrometry analysis
- Computational determination of oxidized/reduced ratios for each cysteine
This approach was recently used to identify 151 proteins with altered redox states in lung fibroblasts from aged mice, with 69% of these changes reversible by Slc7a11 overexpression, highlighting connections between redox regulation and aging [7].
Site-Specific Manipulation of Redox Signaling
Traditional genetic and pharmacological approaches for studying cysteine function face significant limitations. Genetic mutation (e.g., cysteine-to-serine substitution) disrupts all cysteine functions, not just redox sensitivity, while exogenous oxidants lack specificity [6]. Emerging chemical biology strategies address these challenges through precise, site-specific manipulation:
Bioorthogonal Cleavage with Genetic Code Expansion: This approach enables site-specific incorporation of redox-sensitive modifications into proteins of interest. Photocaged cysteine sulfoxide analogs (e.g., DMNB-caged cysteine sulfoxide) can be incorporated as unnatural amino acids via genetic code expansion, allowing controlled generation of sulfenic acid upon UV irradiation [6]. This strategy enables precise temporal and spatial activation of redox signaling at specific sites.
Targeted Covalent Inhibitors (TCIs): Small molecules designed with moderately reactive warheads (e.g., nitroacetamide) can selectively block sulfenic acid modifications at specific target proteins without globally disrupting redox homeostasis [6]. These redox-based TCIs represent a promising therapeutic strategy for diseases involving dysregulated redox signaling.
Table 3: Essential Research Reagents for Studying Cysteine Redox Biology
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Thiol-Alkylating Agents | Iodoacetamide, N-ethylmaleimide (NEM) | Block free thiols to preserve redox states during analysis |
| Chemoselective Probes | Dimedone derivatives (DCP-Bio1, DYn-2) | Selective detection and enrichment of sulfenic acids |
| Redox Biosensors | roGFP, HyPer | Real-time monitoring of redox potential in living cells |
| Mass Tag Reagents | Iodoacetyl TMT, ICAT | Multiplexed quantification of cysteine oxidation states |
| Genetic Tools | CRISPR/Cas9, Redox-sensitive GFP constructs | Manipulation and monitoring of specific redox pathways |
| Low MW Thiol Analogs | Glutathione, Mycothiol, Bacillithiol | Study organism-specific thiol/disulfide systems |
Structural Biology Techniques
High-resolution structural methods including X-ray crystallography and cryo-electron microscopy have provided invaluable insights into the molecular mechanisms of thiol-based redox switches. Structures of redox-sensitive proteins in both reduced and oxidized states have revealed how disulfide formation or sulfenic acid modification triggers conformational changes that alter protein function [2] [4]. For example, structural studies of bacterial OhrR and SarZ have captured sulfenic acid intermediates and documented their structural consequences [2].
Biological Systems and Pathophysiological Implications
Prokaryotic Redox Sensing Systems
Bacteria employ sophisticated thiol-based redox sensors to adapt to oxidative stress encountered in their environments. Among the best-characterized systems is OxyR, a LysR-family transcription factor that senses hydrogen peroxide through disulfide bond formation between conserved cysteine residues [2]. Upon oxidation, OxyR activates expression of a regulon including catalases, peroxidases, and thiol-repair enzymes, coordinating the adaptive response to peroxide stress [2].
Other bacterial redox sensors include:
- OhrR/2-Cys: Repressors that sense organic hydroperoxides through cysteine oxidation
- Spx: A global regulator that controls disulfide stress responses in Gram-positive bacteria
- YodB: A MarR-family repressor that senses various electrophilic stressors
- CrtJ: A photosensor that uses cysteine oxidation to regulate photosynthesis genes
These systems demonstrate the remarkable versatility of cysteine chemistry in environmental sensing and transcriptional regulation [2].
Eukaryotic Redox Signaling in Health and Disease
In eukaryotes, thiol-based redox regulation impacts numerous physiological processes and disease states:
Aging and Longevity: Redox signaling through cysteine modifications has emerged as a key regulator of aging processes. Recent research demonstrates that ROS and hydrogen sulfide (Hâ‚‚S) mediate lifespan extension in model organisms through reversible cysteine oxidation [8] [9]. Proteomic studies reveal that aging is associated with specific changes in the redox states of proteins involved in protein translation, ubiquitin-proteasome function, and cytoskeletal organization, with many of these changes reversible by manipulating redox homeostasis [7].
Fungal Pathogenesis: Pathogenic fungi utilize complex thiol-based systems to survive oxidative stress encountered during infection. Glutathione-dependent enzymes (glutaredoxins, glutathione peroxidases) and thioredoxin systems enable fungal pathogens to neutralize host-derived oxidants and establish infections [10]. These systems represent potential targets for antifungal therapies, particularly as drug resistance becomes increasingly problematic [10].
Metabolic Regulation: Redox-sensitive cysteine residues regulate key metabolic enzymes, allowing metabolic pathways to respond to changes in cellular redox state. For example, oxidation of Cys358 in pyruvate kinase M2 (PKM2) decreases its activity in hyperglycemic conditions, potentially contributing to diabetic complications [6].
The following diagram illustrates a generalized redox signaling pathway from signal perception to physiological response:
The unique chemistry of the cysteine thiol group makes it ideally suited for its role as a cellular redox sensor. The nucleophilicity of the thiolate anion, the diversity of reversible oxidative modifications, and the ability of protein environments to tune cysteine reactivity collectively enable precise sensing of redox challenges and appropriate adaptive responses. As research in this field advances, several emerging areas deserve particular attention:
Chemical Biology Tools: The development of more sophisticated methods for site-specific manipulation of cysteine oxidation states will be crucial for establishing causal relationships between specific redox events and biological outcomes [6]. Bioorthogonal chemistry combined with genetic code expansion represents a particularly promising approach.
Systems-Level Understanding: Integrating redox proteomics with other functional genomics data will provide a more comprehensive view of redox regulation networks and their connections to other cellular processes. This systems biology approach will be essential for understanding how redox signaling becomes dysregulated in disease states.
Therapeutic Targeting: The growing recognition of redox-sensitive cysteine residues as regulatory switches in pathogenic organisms and disease processes highlights their potential as therapeutic targets [10] [6]. Developing targeted covalent inhibitors that specifically modulate redox-sensitive cysteines represents an innovative approach for treating conditions involving oxidative stress.
The study of cysteine-mediated redox signaling continues to reveal the sophisticated mechanisms by which cells perceive and respond to their metabolic and environmental circumstances. As our tools for investigating these processes become increasingly precise, we can anticipate new insights into both fundamental biology and novel therapeutic strategies.
The redox regulation of protein cysteine residues represents a fundamental mechanism in cellular signaling and homeostasis. This in-depth technical guide examines four key oxidative post-translational modifications (oxPTMs) that constitute a critical regulatory network in redox biology: S-sulfenylation, S-nitrosation, S-glutathionylation, and S-persulfidation. These reversible modifications function as molecular sensors that dynamically modulate protein function, structure, and interactions in response to changes in the cellular redox environment [11] [12]. The intricate interplay between these modifications forms a sophisticated signaling language that enables cells to adapt to oxidative stress, regulate metabolic pathways, and control physiological processes ranging from vasodilation to immune response [13] [14]. Disruption of this delicate balance contributes significantly to the pathogenesis of numerous human diseases, including cardiovascular disorders, neurodegeneration, and cancer [11] [15] [14]. This whitepaper provides a comprehensive technical overview of these modifications, focusing on their chemical foundations, detection methodologies, and functional consequences within the broader context of redox regulation research.
Defining the Modifications: Core Concepts and Chemical Profiles
The following table summarizes the core definitions, chemical signatures, and key functional roles of the four cysteine modifications.
Table 1: Fundamental Characteristics of Key Cysteine Oxidative Post-Translational Modifications
| Modification | Chemical Structure | Precursor Species | Key Functional Roles |
|---|---|---|---|
| S-sulfenylation [11] [16] | Cys-SOH | Hydrogen peroxide (Hâ‚‚Oâ‚‚), other ROS [11] | Redox sensor; precursor for other oxPTMs; protects against irreversible oxidation [11] [16] |
| S-nitrosation [17] | Cys-SNO | Nitric oxide (NO), nitrosating agents (e.g., Nâ‚‚Oâ‚„) [18] [17] | Cell signaling (vasodilation, neurotransmission); regulation of kinase/phosphatase activity [17] |
| S-glutathionylation [15] [12] | Cys-S-SG | Oxidized glutathione (GSSG), sulfenic acid intermediate [12] | Protection from oxidative damage; redox regulation of metabolic and signaling proteins [15] [12] |
| S-persulfidation [13] [14] | Cys-SSH | Hydrogen sulfide (Hâ‚‚S), polysulfides (Hâ‚‚Sâ‚™) [13] [14] | Redox signaling; protection against oxidative stress; regulation of metabolism and inflammation [13] [14] [19] |
Formation Pathways and Interrelationships
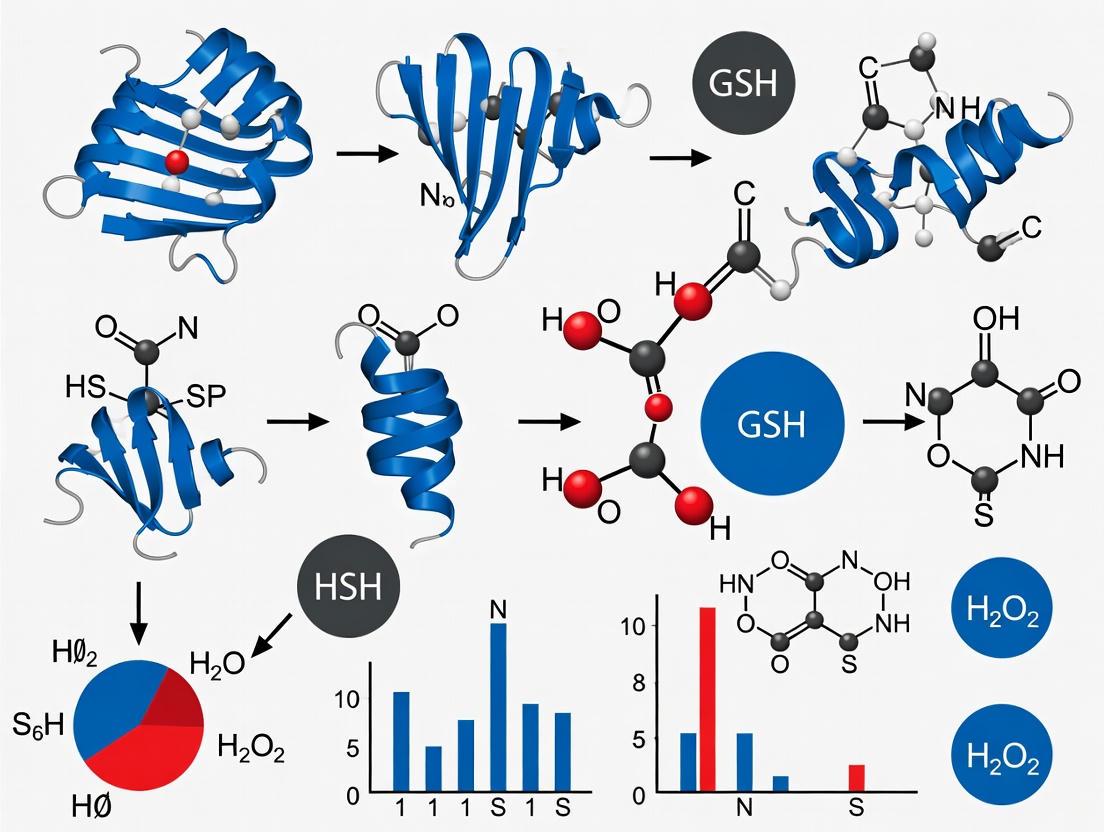
The four modifications do not exist in isolation but are interconnected through shared biochemical pathways. The following diagram illustrates their formation and relationships.
Diagram 1: Cysteine Modification Pathways
S-sulfenylation represents the initial oxidative step, forming a sulfenic acid derivative (Cys-SOH) upon reaction with reactive oxygen species (ROS) like hydrogen peroxide (H₂O₂) [11] [16]. This modification is highly reactive and often transient, serving as a crucial precursor for other oxPTMs. S-glutathionylation occurs when the electrophilic sulfur of a sulfenic acid reacts with the nucleophilic thiol of glutathione (GSH), forming a mixed disulfide [12]. This reaction can protect cysteine residues from irreversible over-oxidation to sulfinic (Cys-SO₂H) or sulfonic (Cys-SO₃H) acids [19]. S-nitrosation involves the attachment of a nitroso group to a cysteine thiol, forming S-nitrosothiols (RSNOs). This can occur through multiple mechanisms, including reaction with nitrous acid or transition metal-catalyzed pathways [17]. S-persulfidation (also called S-sulfhydration) generates a persulfide group (Cys-SSH) on the target protein. This can occur via the reaction of a protein thiol with hydrogen sulfide (H₂S) or, more efficiently, with polysulfides (H₂Sₙ) [13] [14]. Intriguingly, H₂S can also react with S-nitrosothiols to form persulfides, representing a potential cross-talk point between reactive nitrogen and sulfur species [13].
Quantitative Comparison of Modification Characteristics
A thorough understanding of these modifications requires comparing their biochemical properties, stability, and regulatory roles. The following table provides a detailed technical comparison to guide experimental design and data interpretation.
Table 2: Comprehensive Biochemical and Functional Comparison of Cysteine Modifications
| Characteristic | S-sulfenylation | S-nitrosation | S-glutathionylation | S-persulfidation |
|---|---|---|---|---|
| Typical Half-Life | Seconds to minutes [16] | Minutes to hours | Minutes to hours [12] | Minutes [13] |
| Key Detection Methods | Dimedone probes [20] [16], DYn-2 [20] | Biotin-switch technique, Saville-Griess assay | Anti-GSH antibodies, biotinylated GSH esters [12] | Tag-switch assay, SSP4 probe [13] [14] |
| Reversibility | Highly reversible | Reversible | Reversible [12] | Highly reversible |
| Primary Regulatory Role | Early oxidative stress sensor | Signaling molecule in vasodilation, neurotransmission [17] | Protection from over-oxidation, redox regulation [12] | Hâ‚‚S-mediated signaling, oxidative stress defense [13] |
| Enzymatic Reversal Systems | Thioredoxin, Glutaredoxin (via reduction of disulfides) | Thioredoxin, S-nitrosoglutathione reductase | Glutaredoxin, Sulfiredoxin | Thioredoxin, Sulfiredoxin [13] |
| Estimated Abundance | 5-12% of cysteines oxidized (normal conditions) [11] | Tissue and protein-specific | Significant during oxidative stress | 10-25% of liver proteome [13] |
Experimental Protocols and Research Toolkit
Accurate detection of these labile modifications presents significant technical challenges. This section outlines key experimental workflows and reagents essential for rigorous research in this field.
Key Detection Workflows
Protocol for S-Sulfenylation Detection Using Chemical Probes
The most reliable method for detecting protein S-sulfenylation utilizes nucleophilic probes like dimedone that selectively trap the sulfenic acid intermediate. The following workflow is adapted from high-throughput proteomic studies [20].
- Cell Lysis and Blocking: Lyse cells under non-reducing conditions in the presence of alkylating agents such as iodoacetamide (IAM) or N-ethylmaleimide (NEM) to block free thiols and prevent post-lysis oxidation.
- Probe Incubation: Incubate the lysate with a sulfenic acid-specific probe (e.g., dimedone or its biotin-conjugated derivative DCP-Bio1) for 1-2 hours.
- Streptavidin Pulldown: If a biotinylated probe is used, capture the labeled proteins using streptavidin-coated beads.
- Washing and Elution: Wash beads stringently to remove non-specifically bound proteins. Elute the modified proteins for analysis.
- Downstream Analysis: Identify and quantify the modified proteins using techniques such as western blotting (for known targets) or mass spectrometry (for proteomic profiling) [20].
Protocol for S-Persulfidation Detection via Tag-Switch Assay
The tag-switch assay is a widely used method to selectively label protein persulfides, distinguishing them from other modifications like glutathionylation [13].
- Free Thiol Blocking: Treat the protein sample with methyl methanethiosulfonate (MMTS) to block all free thiols (Cys-SH) and other thiol-based modifications. MMTS also reacts with persulfides (Cys-SSH) to form Cys-S-S-CH₃.
- Selective Reduction: The key step involves treating the sample with a phosphine-based reducing agent (e.g., tris(2-carboxyethyl)phosphine, TCEP) that selectively reduces the Cys-S-S-CH₃ derivative back to a free persulfide (Cys-SSH), but does not reduce disulfide bonds like those in S-glutathionylation.
- Labeling: The newly regenerated persulfide thiol is then labeled with a biotin- or fluorophore-tagged alkylating agent (e.g., biotin-maleimide).
- Detection: The labeled persulfidated proteins can be detected and analyzed via western blot, mass spectrometry, or other appropriate methods.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for Studying Cysteine Redox Modifications
| Reagent / Tool | Chemical Nature | Primary Function in Research |
|---|---|---|
| Dimedone & DCP-Bio1 [20] [16] | 5,5-dimethyl-1,3-cyclohexanedione (and biotin conjugate) | Selective chemical probe that covalently labels and traps protein sulfenic acids (-SOH) for detection. |
| MMTS [13] [19] | Methyl Methanethiosulfonate | Small membrane-permeable alkylating agent used to block free thiols and as a key reagent in the tag-switch assay for persulfidation. |
| Biotin-HPDP | N-(6-(Biotinamido)hexyl)-3'-(2'-pyridyldithio)propionamide | Thiol-reactive biotinylation reagent used in techniques like the biotin-switch assay for S-nitrosation. |
| GYY4137 [14] | Morpholin-4-ium 4 methoxyphenyl(morpholino) phosphinodithioate | Slow-releasing hydrogen sulfide (Hâ‚‚S) donor used to study the biological effects of Hâ‚‚S and induce protein persulfidation. |
| Sodium Nitroprusside [17] | Nitrosyl pentacyanoferrate(III) | Nitric oxide (NO) donor used in experimental settings to induce protein S-nitrosation. |
| AP39 [14] | Mitochondria-targeted Hâ‚‚S donor | A [10-oxo-10-(4-thiocyanatophenyl)decyl]triphenylphosphonium derivative that delivers Hâ‚‚S specifically to mitochondria. |
| 2-Amino-3-iodopyridine | 2-Amino-3-iodopyridine, CAS:104830-06-0, MF:C5H5IN2, MW:220.01 g/mol | Chemical Reagent |
| 4-Bromo-2-fluoro-1-iodobenzene | 4-Bromo-2-fluoro-1-iodobenzene, CAS:105931-73-5, MF:C6H3BrFI, MW:300.89 g/mol | Chemical Reagent |
Biological Roles and Implications for Drug Development
These cysteine modifications regulate critical cellular processes, and their dysregulation is implicated in major human diseases, making them attractive targets for therapeutic intervention.
Functional Consequences and Disease Links
S-sulfenylation in Redox Sensing and Stability: S-sulfenylation acts as a protective switch. By reversibly forming sulfenic acid, cysteine residues can be shielded from irreversible oxidation to sulfinic or sulfonic acids, which would lead to permanent protein damage [11] [16]. This modification also directly regulates protein function; for example, it can inhibit the activity of protein tyrosine phosphatases (PTPs), thereby enhancing kinase signaling [11]. Its role is emerging in thrombotic disorders, diabetes, cardiovascular diseases, neurodegenerative diseases, and cancer [11].
S-nitrosation in Cell Signaling: S-nitrosation is a key mediator of nitric oxide (NO) signaling, regulating a wide range of physiological processes including vasodilation, neurotransmission, and immune response [17]. It exerts regulatory control over many enzymes, typically suppressing the activity of kinases like JNK and IKKβ, while also inhibiting protein tyrosine phosphatases (PTPs) [17]. This modification is crucial in cardiovascular and neuronal systems, with alterations observed in conditions like heart failure and neurodegeneration.
S-glutathionylation in Metabolic Regulation and Cancer: This modification provides a mechanism for the redox regulation of central metabolic enzymes, effectively linking the cellular redox state to metabolic flux [15] [12]. It plays a vital protective role by shielding cysteines from irreversible oxidation during oxidative stress [12]. In cancer, altered S-glutathionylation patterns are linked to drug resistance, making the enzymes responsible for its reversal (e.g., glutaredoxin) potential therapeutic targets [15].
S-persulfidation in Physiology and Disease: Persulfidation is increasingly recognized as a fundamental redox regulation mechanism. It reprograms cysteine sensors in critical pathways involving metabolism (e.g., GAPDH), inflammation (e.g., NLRP3), and transcription (e.g., Keap1/NRF2) [14]. It has been shown to augment GAPDH activity and enhance actin polymerization, directly influencing cellular physiology [13]. Disrupted persulfidation is implicated in cardiovascular diseases, neurodegenerative disorders, metabolic syndromes, and cancer [14]. Endogenously produced Hâ‚‚S and subsequent persulfidation have been demonstrated as critical for physiological functions such as sperm viability [19].
Therapeutic Targeting and Future Directions
The strategic manipulation of cysteine oxPTMs represents a promising frontier in pharmacology. Several therapeutic approaches are under investigation:
- Hâ‚‚S Donors: Slow-releasing Hâ‚‚S donors like GYY4137 and mitochondria-targeted donors like AP39 are being explored for their ability to restore protective persulfidation signaling in cardiovascular disease, inflammation, and metabolic disorders [14]. The donor SG1002 has progressed to Phase II clinical trials (NCT02278276) [14].
- Diet-Derived Compounds: Natural polysulfides from garlic and isothiocyanates from cruciferous vegetables can modulate the cellular persulfidome and activate cytoprotective pathways like Nrf2, offering potential for nutritional interventions [14].
- Enzyme Inhibitors: Developing inhibitors or activators of the specific enzymes that write, erase, or read these modifications (e.g., CSE/CBS for Hâ‚‚S production, glutaredoxin for deglutathionylation) is a key strategy [15] [14].
A major challenge in the field is the narrow therapeutic margin of many donors and the difficulty in real-time quantification of these modifications in vivo. Future efforts will focus on developing smarter, tissue-targeted reagents and combining them with advanced detection techniques to translate our understanding of redox signaling into effective therapies.
The redox regulation of protein cysteine residues represents a fundamental mechanism in cellular signaling and homeostasis. Cysteine (Cys), a sulfur-containing amino acid, serves as a pivotal player in cellular redox regulation through its highly reactive thiol (–SH) group [21]. This thiol group is subject to a variety of posttranslational modifications (PTMs), such as sulfenylation, sulfinylation, glutathionylation, and nitrosylation, which modulate protein functions, redox homeostasis, and cellular responses to stress [21]. The dynamic nature of these modifications allows cells to rapidly respond to changing oxidative conditions, making cysteine-based redox switches crucial for adapting to physiological and pathological challenges. Dysregulation of these Cys-based PTMs has been implicated in disease progression, making them potential targets for therapeutic intervention [21]. Within this framework, three key regulatory systems have emerged as central players: the peroxiredoxin (Prx) family of peroxidases, the thioredoxin (Trx) system that maintains them in a reduced state, and sulfiredoxin-1 (SRXN1), which serves a unique repair function for oxidized Prxs [21] [22] [23].
Molecular Properties and Classification
Peroxiredoxins: Ubiquitous Thiol-Peroxidases
Peroxiredoxins (Prxs) are a fascinating group of thiol-dependent peroxidases (EC 1.11.1.15) that detoxify Hâ‚‚Oâ‚‚, aliphatic and aromatic hydroperoxides, and peroxynitrite [22]. They are ubiquitously expressed, with multiple isoforms present in most organisms (e.g., 3 isoforms in Escherichia coli, 5 in Saccharomyces cerevisiae, 6 in Homo sapiens, and 9 in Arabidopsis thaliana) [22]. The classification of Prxs has evolved from initial mechanistic distinctions to more sophisticated bioinformatics approaches:
Table 1: Peroxiredoxin Classification and Properties
| Classification | Representative Members | Conserved Cysteines | Structural Features | Reducing Partner |
|---|---|---|---|---|
| Typical 2-Cys Prx | PrxI-IV (mammals), Tsa1 (yeast), AhpC (bacteria) | Cp and CR | Homodimeric with intersubunit disulfide | Thioredoxin |
| Atypical 2-Cys Prx | PrxV (mammals) | Cp and CR | Intrasubunit disulfide | Thioredoxin |
| 1-Cys Prx | PrxVI (mammals) | Cp only | No resolving cysteine | Glutathione, Ascorbate? |
The absolutely conserved peroxidatic cysteine (C~P~) residue serves as the site of oxidation by peroxides [23]. Mammalian cells express six Prx isoforms (PrxI to PrxVI), which are classified into the three groups above based on the location or absence of the resolving cysteine (C~R~) [23]. A more recent "global evolutionary classification" divided Prx proteins into six subfamilies: Prx1, Prx5, Prx6, Tpx, PrxQ, and AhpE, with mammalian PrxI-IV belonging to Prx1, PrxV to Prx5, and PrxVI to Prx6 [24].
The Thioredoxin System: Central Redox Hub
The thioredoxin (TRX) system is one of the major cellular antioxidant pathways that control redox homeostasis [25]. This system comprises NADPH, TRX reductase (TRXR), TRX itself, and the negative regulator TRX-interacting protein (TXNIP) [25]. TRXR is a selenoenzyme that utilizes reducing equivalents from NADPH generated by the pentose phosphate pathway (PPP) to keep TRX in its reduced state [25]. Mammals possess three TRXR isoforms with distinct subcellular localizations: TRXR1 (cytoplasm), TRXR2 (mitochondria), and TRXR3 (testis-specific), along with corresponding TRX isoforms (TRX1 and TRX2) [25].
Thioredoxin is a 12-kD oxidoreductase protein with a characteristic tertiary structure termed the thioredoxin fold [26]. The active site contains a dithiol in a CXXC motif that is essential for its reductase activity [26]. The related glutaredoxins share many thioredoxin functions but are reduced by glutathione rather than a specific reductase, creating parallel but interconnected redox systems [26].
Sulfiredoxin-1: Redox Repair Enzyme
Sulfiredoxin-1 (SRXN1) is an essential component of the cellular redox system, specifically tasked with reversing the hyperoxidation of peroxiredoxins (Prxs), thereby restoring their peroxidase activity [21]. Human SRXN1 consists of 137 amino acids and has a molecular weight of approximately 14 kDa [21]. The Cys residue at position 99 (Cys99) in SRXN1 is directly involved in the enzymatic mechanism that facilitates the reduction of sulfinic acid, and mutations at this Cys abolish desulfinylation activity [21]. SRXN1 catalyzes the ATP-dependent reduction of Cys sulfinic acid (Cys-SOâ‚‚H) within hyperoxidized Prxs, a previously unknown reaction in biological systems [21].
Table 2: Key Redox Regulators: Genetic and Biochemical Properties
| Redox Component | Human Gene | Protein Size | Key Structural Motifs | Cellular Localization |
|---|---|---|---|---|
| SRXN1 | SRXN1 | 14 kDa (137 aa) | Critical Cys99 | Cytosol |
| Typical 2-Cys Prx | PRDX1-PRDX4 | ~22 kDa per subunit | PxxxTxxC~P~ motif | Cytosol, Mitochondria, Nucleus, Secretory Pathway |
| Thioredoxin 1 | TXN | 12 kDa | CXXC active site motif | Cytoplasm |
| Thioredoxin Reductase 1 | TXNRD1 | ~55 kDa subunit | Selenocysteine residue | Cytoplasm |
Catalytic Mechanisms and Functional Roles
The Peroxiredoxin Catalytic Cycle
The catalytic cycle of Prxs begins with the nucleophilic attack of the peroxidatic cysteine (C~P~-Sâ») on the peroxide substrate, resulting in the formation of cysteine sulfenic acid (C~P~-SOH) and the release of water or alcohol [22] [23]. The C~P~ of Prx is oxidized by peroxides very rapidly with second-order rate constants of 10â¶-10⸠Mâ»Â¹sâ»Â¹, which are 5-7 orders of magnitude higher than those for small thiols [23]. This exceptional reactivity is enabled by a conserved transition state involving an extensive hydrogen bond network that comprises the C~P~ thiolate anion, peroxide, Thr and Pro residues within a universal PXXXTXXC~P~ active site motif, and a conserved Arg residue [23].
In the next step, resolution of the sulfenic acid intermediate occurs through disulfide bond formation. For typical 2-Cys Prxs, the sulfenic acid reacts with the resolving cysteine (C~R~-SH) from the other subunit to form an intersubunit disulfide bond [22]. For atypical 2-Cys Prxs like PrxV, this disulfide forms within the same subunit [23]. This resolution step requires a localized unfolding of structures around the C~P~ that removes the S~P~OH from the "fully folded" (FF) active site and exposes it in the "locally unfolded" (LU) form for disulfide bond formation [22]. The disulfide form is then reduced by thioredoxin to complete the catalytic cycle [22].
Thioredoxin System Redox Transfer
The thioredoxin system operates as a redox relay that transfers reducing equivalents from NADPH to target proteins [25]. The process begins with TRXR utilizing NADPH to reduce its FAD cofactor, which then reduces the active site disulfide in TRX [26]. Reduced TRX, with its CXXC motif in the dithiol state, can then reduce disulfide bonds in target proteins such as Prxs [26]. For Trx1, this process involves attack of Cys32 on the oxidized group of the substrate, followed immediately by disulfide bond formation with Cys35, thereby transferring two electrons to the substrate [26]. Oxidized Trx1 is then reduced by thioredoxin reductase, completing the cycle [26].
Sulfiredoxin-1-Mediated Repair of Hyperoxidized Peroxiredoxins
During their catalytic cycle, particularly under conditions of high oxidative stress, the sulfenic acid intermediate (C~P~-SOH) of Prxs can be further oxidized to sulfinic acid (C~P~-SOâ‚‚H), leading to irreversible inactivation [21]. SRXN1 catalyzes the ATP-dependent reduction of this sulfinic acid modification, restoring the catalytic cysteine to its reduced state [21]. The mechanism involves a unique ATP-dependent process where SRXN1 forms a covalent intermediate with the oxidized Prx, followed by thiol-mediated reduction [21]. This repair function is specific to 2-Cys peroxiredoxins and represents a critical defense mechanism against oxidative damage [21].
Figure 1: Integrated Redox Cycles of Peroxiredoxins, Thioredoxin, and Sulfiredoxin-1. This diagram illustrates the catalytic cycle of peroxiredoxins (Prx) in peroxide reduction, their regeneration by the thioredoxin (Trx) system, and the repair of hyperoxidized Prx by sulfiredoxin-1 (SRXN1).
Redox Signaling in Cellular Processes
Regulation of Immune Cell Function
The TRX system plays a particularly important role in immune cell activation and function [25]. Upon T cell activation, the TRX1-TRXR1 system is upregulated and supports massive proliferation by providing reducing equivalents to ribonucleotide reductase (RNR), which is essential for deoxyribonucleotide (dNTP) production during DNA biosynthesis [25]. Genetic deletion of Txnrd1 in mice results in impaired expansion of activated T cells during viral infections, demonstrating its critical role in adaptive immunity [25]. Interestingly, Txnrd1-deficient T cells do not display increased levels of ROS, indicating that the primary defect is in nucleotide synthesis rather than antioxidant defense [25].
B cells utilize a more flexible redox system, with the ability to employ glutaredoxin 1 (GRX1) as a compensatory pathway when TRX is unavailable [25]. However, innate-like B cells (B1 and marginal zone B cells) resemble T cells in their strict requirement for the TRX system, as they are unable to engage GRX1 compensation [25]. This differential requirement highlights the cell type-specific adaptations in redox regulation.
Redox Control of Gene Expression
The TRX system regulates several transcription factors, including NF-κB, AP-1, HIF1α, and the antioxidant master regulator Nrf2 [25] [26]. TRX1 reduces a disulfide bond in NF-κB, promoting its DNA-binding activity [26]. Similarly, TRX1 indirectly increases AP-1 DNA-binding activity by reducing the DNA repair enzyme redox factor 1 (Ref-1), which in turn reduces AP-1 [26]. This creates a redox regulation cascade that fine-tunes gene expression in response to oxidative stimuli.
SRXN1 itself is transcriptionally regulated by both Nrf2 and AP-1, creating feedback loops that enhance cellular antioxidant capacity [21]. The SRXN1 promoter contains a functional antioxidant response element (ARE) at -228 bp from the transcription start site that is responsive to Nrf2, as well as AP-1 binding sites that mediate induction in response to other stimuli [21].
Role in Cell Survival and Death Decisions
Redox regulators influence critical decisions between cell survival and death through multiple mechanisms. TRX1 regulates apoptosis by suppressing ASK1 activity through direct binding [25]. Under normal conditions, reduced TRX1 binds to ASK1 and inhibits its kinase activity. During oxidative stress, TRX1 is oxidized and dissociates from ASK1, allowing ASK1 to activate JNK and p38 MAPK pathways that promote apoptosis [25].
Peroxiredoxins also participate in cell death decisions through their chaperone activity. Upon hyperoxidation, some Prxs can form high molecular weight complexes that gain molecular chaperone function, protecting proteins from aggregation under stress conditions [24]. This switch from peroxidase to chaperone function represents an adaptive response to oxidative stress.
Experimental Approaches and Methodologies
Assessing Redox Status and Enzyme Activity
Table 3: Key Methodologies for Studying Redox Regulators
| Methodology | Application | Key Insights Provided |
|---|---|---|
| Competitive kinetics with HRP or catalase | Measuring Prx reaction rates | Second-order rate constants of 10â¶-10⸠Mâ»Â¹sâ»Â¹ for peroxide reduction |
| Stopped-flow fluorescence | Monitoring reaction intermediates | Identification of sulfenic acid formation and resolution kinetics |
| Site-directed mutagenesis | Functional analysis of catalytic residues | Cys99 in SRXN1 and C~P~ in Prxs are essential for activity |
| Redox-sensitive GFP probes | Compartment-specific redox measurements | Spatial organization of redox signaling |
| Thioredoxin trapping mutants | Identifying TRX substrates | CXXC motif mutants trap intermediate disulfides |
Transcriptional Regulation Studies
The transcriptional regulation of SRXN1 has been elucidated through promoter analysis techniques. Deletion constructs and site-directed mutagenesis of the SRXN1 promoter identified functional ARE and AP-1 binding sites [21]. Chromatin immunoprecipitation (ChIP) assays confirmed the binding of Nrf2 and AP-1 transcription factors to these regulatory elements [21]. Luciferase reporter assays demonstrated that Nrf2 knockdown decreases SRXN1 promoter activity, while Nrf2 activators like tBHQ significantly enhance its activity [21].
Genetic Manipulation Approaches
Genetic studies including knockout mouse models have been instrumental in defining the physiological functions of redox regulators. Mice lacking peroxiredoxin 1 or 2 develop severe hemolytic anemia and are predisposed to certain hematopoietic cancers, while Peroxiredoxin 1 knockout mice have a 15% reduction in lifespan [24]. Tissue-specific knockout approaches, such as Txnrd1 deletion in T cells, have revealed cell-type-specific requirements for redox enzymes [25]. Similarly, CRISPR-Cas9 screens identified Txnrd1 as a positive regulator of the antitumor response of CD8+ T cells [25].
Figure 2: Transcriptional Regulation of SRXN1 and its Functional Consequences. This diagram illustrates how oxidative stress activates transcription factors Nrf2 and AP-1, which bind to antioxidant response elements (ARE) in the SRXN1 promoter to increase SRXN1 expression, ultimately leading to repair of hyperoxidized peroxiredoxins and restoration of redox homeostasis.
Pathophysiological Implications and Therapeutic Targeting
Role in Liver Disease
SRXN1 has emerged as a significant factor in liver pathophysiology [21]. Through its regulation of Cys sulfinylation across a broad spectrum of liver diseases, SRXN1 modulates redox-sensitive signaling pathways that govern inflammation, apoptosis, and cell survival [21]. The critical role of SRXN1 in regulating oxidative stress and cellular signaling through its interaction and desulfinylation of target proteins is crucial to maintaining cellular function under pathological conditions [21]. A deeper understanding of SRXN1-mediated redox regulation may offer a novel therapeutic avenue to mitigate Cys oxidation and improve clinical outcomes in various liver disease contexts [21].
Connections to Cancer and Aging
Redox regulators display dual roles in cancer, functioning as both tumor suppressors and promoters depending on context [21] [25]. The TRX system is considered a promising target in cancer therapy, with inhibitors undergoing clinical evaluation [25]. Similarly, SRXN1 has been implicated in cancer progression, with studies showing it promotes hepatocellular carcinoma growth by inhibiting TFEB-mediated autophagy and lysosome biogenesis [27]. SRXN1 also stimulates hepatocellular carcinoma tumorigenesis and metastasis through modulating ROS/p65/BTG2 signaling [27].
The thioredoxin system has been directly linked to cellular senescence and aging-related diseases [28]. The progressive decline of redox homeostasis with age contributes to the pathogenesis of multiple age-related conditions, making redox regulators potential targets for therapeutic intervention in aging [28].
Therapeutic Targeting Strategies
Several strategies have emerged for targeting redox regulators therapeutically:
TRX System Inhibitors: Compounds like auranofin inhibit thioredoxin reductase and have shown promise in cancer therapy by disrupting redox balance in rapidly dividing cells [25].
Nrf2 Activators: Since SRXN1 is transcriptionally regulated by Nrf2, compounds that activate Nrf2 signaling could enhance cellular antioxidant defenses in degenerative diseases [21].
SRXN1 Expression Modulation: Both upregulation and inhibition of SRXN1 may have therapeutic value depending on context, with SRXN1 inhibition potentially sensitizing cancer cells to oxidative stress-inducing therapies [27].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Studying Redox Regulation
| Reagent/Category | Specific Examples | Research Application | Function in Experiments |
|---|---|---|---|
| Expression Plasmids | SRXN1, PRDX1-6, TXN, TXNRD1 | Overexpression studies | Functional analysis of wild-type vs. mutant proteins |
| Mutant Constructs | C99S SRXN1, C~P~ mutants of Prxs | Structure-function studies | Determining essential catalytic residues |
| Knockout Models | Global and tissue-specific KO mice | Physiological studies | Defining essential functions in different tissues |
| Specific Inhibitors | Auranofin (TRXR inhibitor) | Therapeutic targeting studies | Evaluating consequences of system inhibition |
| Activity Probes | Redox-sensitive fluorescent dyes | Real-time monitoring | Visualizing compartment-specific redox changes |
| Antibodies | Anti-SRXN1, anti-Prx-SOâ‚‚H | Detection in cells and tissues | Assessing expression and oxidation status |
| Conduritol B Tetraacetate | Conduritol B Tetraacetate|CAS 25348-63-4 | Bench Chemicals | |
| Aleoe-emodin triacetate | Triacetyl Aloe-emodin | 1,8-Bis(acetyloxy)-3-[(acetyloxy)methyl]-9,10-anthracenedione | Bench Chemicals |
Future Perspectives and Research Directions
The field of cysteine redox regulation continues to evolve with several emerging research directions. The development of more specific tools to monitor and manipulate individual redox systems in specific cellular compartments remains a priority. Understanding the complex interactions and potential redundancies between the TRX, glutathione, and other redox systems in different cell types and disease contexts represents another important frontier. The therapeutic targeting of these systems requires greater specificity to avoid disrupting essential redox homeostasis while achieving desired therapeutic effects.
Single-cell analysis of redox states and the integration of redox proteomics with other omics approaches will likely provide unprecedented insights into the spatial and temporal organization of redox signaling networks. Furthermore, the role of redox regulation in emerging areas such as the microbiome-brain axis and immunometabolism represents fertile ground for future investigation.
In conclusion, Sulfiredoxin-1, peroxiredoxins, and the thioredoxin system represent interconnected components of the cellular redox regulatory network that fine-tune cysteine-based signaling events. Their study continues to yield fundamental insights into cellular homeostasis and provides promising avenues for therapeutic intervention in diverse disease contexts.
The cellular antioxidant response is a critical defense mechanism against oxidative stress, a pervasive challenge in both physiological homeostasis and pathological states. This whitepaper delineates the sophisticated transcriptional machinery governed by the transcription factors Nrf2 and AP-1, which coordinately regulate a vast network of cytoprotective genes. Central to this discussion is their synergistic and context-dependent regulation of target genes, such as sulfiredoxin (SRXN1), through shared cis-regulatory elements. Framed within the broader context of redox regulation of protein cysteine residues, this review integrates current mechanistic insights, experimental methodologies, and therapeutic implications. The content is specifically tailored for researchers, scientists, and drug development professionals seeking a deeper understanding of these pathways for therapeutic intervention in diseases characterized by oxidative damage, including neurodegenerative disorders and cancer.
Oxidative stress, characterized by an imbalance between the production of reactive oxygen species (ROS) and the cellular antioxidant defenses, is a well-established contributor to the pathogenesis of a wide array of diseases [29] [30]. The nervous system is particularly vulnerable, and oxidative stress is a significant factor in neurodegenerative diseases such as Parkinson's disease (PD) and amyotrophic lateral sclerosis (ALS) [30]. At a molecular level, the thiol group of cysteine residues in proteins is a primary target of ROS, leading to a spectrum of post-translational modifications (PTMs)—including sulfinylation, glutathionylation, and nitrosylation—that profoundly influence protein function, redox homeostasis, and cellular signaling [29].
To manage this constant threat, cells have evolved intricate transcriptional programs that enhance the expression of antioxidant and detoxification enzymes. The transcription factors Nuclear Factor Erythroid 2-Related Factor 2 (Nrf2) and Activator Protein-1 (AP-1) stand as pivotal regulators of this inducible defense system [31] [32]. This review will explore their unique and overlapping roles, the molecular mechanisms of their activation, and their coordinated regulation of a shared genetic battery, with a specific focus on the redox regulator sulfiredoxin-1.
The Nrf2-Keap1 Signaling Axis
Molecular Architecture and Basal Regulation
Nrf2 is a member of the Cap 'n' Collar (CNC) subfamily of basic-region leucine zipper (bZIP) transcription factors [33]. Its activity is tightly controlled by its cytoplasmic repressor, Kelch-like ECH-associated protein 1 (Keap1). Under homeostatic conditions, Nrf2 is constitutively targeted for ubiquitination and proteasomal degradation. This process is facilitated by Keap1, which acts as a substrate adaptor for a Cullin 3 (Cul3)-based E3 ubiquitin ligase complex. Nrf2 is sequestered in the cytoplasm via binding through its two motifs, the ETGE and DLG motifs, which interact with the Kelch domain of Keap1 [33]. This arrangement ensures a rapid turnover of Nrf2, maintaining low basal levels under non-stressed conditions.
Mechanism of Activation by Inducers
The activation of Nrf2 is primarily post-translational and hinges on the modification of critical cysteine residues within Keap1 and Nrf2 itself. Keap1 contains numerous cysteine residues that function as redox sensors. Specific inducers, including the isothiocyanate sulforaphane (SFN) and tert-butylhydroquinone (tBHQ), modify key cysteine residues (e.g., C151 in the BTB domain and C273 and C288 in the linker region) leading to a conformational change in Keap1 [34] [35]. This modification disrupts the Keap1-Cul3 E3 ligase activity, impairing Nrf2 ubiquitination. Consequently, newly synthesized Nrf2 escapes degradation, stabilizes, and translocates to the nucleus [33] [35].
Recent evidence also underscores the direct role of Nrf2 cysteine residues. Mutation of critical cysteines in Nrf2 (Cys235, Cys311, Cys316, Cys414, and Cys506) enhances its binding to Keap1, increases its polyubiquitination, and shortens its half-life. Furthermore, mutations at Cys119, Cys235, and Cys506 can impede the binding of Nrf2 to the Antioxidant Response Element (ARE) and its recruitment of the coactivator CBP/p300, illustrating that Nrf2 itself is a redox-sensitive protein [34].
Transcriptional Programming via the ARE
Once in the nucleus, Nrf2 heterodimerizes with small Maf (sMaf) proteins and binds to the cis-acting Antioxidant Response Element (ARE), with a core sequence of 5'-TGACXXXGC-3', in the promoter regions of its target genes [33]. This program coordinates the expression of over 200 genes involved in glutathione biosynthesis (e.g., GCLC, GCLM), ROS detoxification (e.g., NQO1, HO-1), xenobiotic metabolism, and NADPH regeneration [33]. The loss of Nrf2 in knockout mice leads to increased susceptibility to various chemical toxicities, autoimmune dysfunction, and cancer, highlighting its non-redundant role as a "guardian of redox homeostasis" [34] [33].
The AP-1 Signaling Pathway
Composition and Regulation
AP-1 is not a single transcription factor but a collection of dimeric complexes composed of members from the Jun (c-Jun, JunB, JunD), Fos (c-Fos, FosB, Fra-1, Fra-2), ATF, and Maf subfamilies [29] [32]. These proteins contain a basic leucine zipper (bZIP) domain that mediates dimerization and DNA binding. The composition of the AP-1 dimer determines its binding affinity and transcriptional activity [32]. AP-1 activity is rapidly induced by a diverse array of stimuli, including growth factors, cytokines, and cellular stresses. Its regulation is complex, occurring at both transcriptional and post-translational levels, with kinases from the MAPK pathways (JNK, ERK, p38) playing a central role in phosphorylating and activating components like c-Jun [32].
Genomic Targets and Functional Diversity
AP-1 dimers recognize either the TPA response element (TRE; 5'-TGAG/CTCA-3') or the cAMP response element (CRE) to regulate genes involved in cell proliferation, differentiation, apoptosis, and inflammation [32]. Its role in the antioxidant response is context-dependent. While AP-1 can drive pro-survival gene expression, it can also contribute to tumor promotion and inflammatory processes. The functional outcome of AP-1 activation is thus highly specific to the cellular context, dimer composition, and nature of the stimulus.
Integrated Regulation: The Sulfiredoxin (SRXN1) Case Study
The gene encoding Sulfiredoxin-1 (SRXN1) serves as a paradigm for the integrated regulation of the antioxidant response by Nrf2 and AP-1, bridging the gap between transcriptional control and cysteine redox regulation.
Functional Role of SRXN1: SRXN1 is a key redox repair enzyme that catalyzes the ATP-dependent reduction of cysteine-sulfinic acid (Cys-SO2H) on hyperoxidized Peroxiredoxins (Prxs), thereby reactivating these critical antioxidant enzymes [31] [29]. This activity places SRXN1 at the heart of the cellular machinery that reverses oxidative cysteine modifications.
Promoter Architecture: The promoter of the human
SRXN1gene contains a conserved ARE. Notably, the core ARE sequence embeds a proximal AP-1 binding site, a common feature in many Nrf2 target genes [31] [36]. This physical overlap suggests a mechanism for synergistic or competitive interactions.Dual Transcriptional Control:
- Nrf2-dependent Regulation: SRXN1 has been conclusively identified as an Nrf2 target gene. The primary functional ARE (ARE1) is located at -228 bp from the transcription start site. Nrf2 activators like tBHQ and synthetic triterpenoids (e.g., CDDO-TFEA) potently induce
SRXN1transcription, and this induction is abrogated upon Nrf2 knockdown [31] [29] [36]. - AP-1-dependent Regulation: AP-1 also regulates
SRXN1expression. For instance, in neurons, synaptic activity inducesSRXN1via AP-1, conferring protection against oxidative stress [31] [36]. The specific composition of the AP-1 dimer (e.g., Jun–Jun or Jun–Fos) can influence the transcriptional output from the shared ARE/AP-1 element.
- Nrf2-dependent Regulation: SRXN1 has been conclusively identified as an Nrf2 target gene. The primary functional ARE (ARE1) is located at -228 bp from the transcription start site. Nrf2 activators like tBHQ and synthetic triterpenoids (e.g., CDDO-TFEA) potently induce
Biological and Therapeutic Significance: The Nrf2-mediated induction of SRXN1 is a crucial part of the protective response in models of hyperoxic lung injury [31]. Furthermore, in liver diseases, SRXN1 has been shown to modulate redox-sensitive signaling pathways governing inflammation, apoptosis, and cell survival, making it a potential therapeutic target [29]. This dual regulation allows the cell to fine-tune the expression of a critical repair enzyme through multiple signaling pathways, ensuring a robust defense against various oxidative insults.
Experimental Methods for Studying Nrf2 and AP-1
To investigate the complex regulation of Nrf2 and AP-1, researchers employ a multifaceted toolkit. The table below summarizes key experimental protocols and the reagents used to dissect these pathways.
Table 1: Key Experimental Protocols for Nrf2 and AP-1 Research
| Methodology | Key Steps | Application in Nrf2/AP-1 Research |
|---|---|---|
| Luciferase Reporter Assay [29] [32] | 1. Clone promoter region (e.g., SRXN1 or synthetic ARE) into luciferase vector.2. Co-transfect with Nrf2/AP-1 expression plasmids.3. Treat with compounds (e.g., SFN, EGCG).4. Measure luciferase activity. |
Quantifies transcriptional activity and promoter binding; identifies functional ARE/AP-1 elements and assesses effects of pharmacological activators/inhibitors. |
| Chromatin Immunoprecipitation (ChIP) [34] | 1. Cross-link proteins to DNA.2. Shear chromatin.3. Immunoprecipitate with antibody against Nrf2 or AP-1 components.4. Reverse cross-links and quantify bound DNA via PCR. | Confirms direct, physical binding of transcription factors to specific genomic regions (e.g., the SRXN1 promoter). |
| Gene Expression Analysis (qRT-PCR) [37] | 1. Treat cells (e.g., SH-SY5Y) with inducer.2. Extract total RNA and synthesize cDNA.3. Perform quantitative PCR with primers for NQO1, HO-1, SRXN1. |
Measures endogenous mRNA levels of target genes to confirm pathway activation. |
| Protein Stability & Ubiquitination Assays [34] [35] | 1. Treat cells with proteasome inhibitor (MG132).2. Immunoprecipitate Nrf2.3. Immunoblot with anti-ubiquitin antibody. | Demonstrates Keap1-dependent ubiquitination and stabilizes Nrf2 for detection. |
| Cysteine Mutagenesis [34] | 1. Generate Nrf2 or Keap1 mutants with Cys-to-Ala substitutions.2. Express in knockout cells (e.g., Nrf2-KO MEFs).3. Assess protein half-life, Keap1 binding, and transcriptional activity. |
Identifies critical cysteine residues for chemical sensing, protein-protein interactions, and degradation. |
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Investigating Nrf2 and AP-1 Pathways
| Reagent / Tool | Function & Utility |
|---|---|
| Cell Lines | |
Nrf2-Knockout (KO) Mouse Embryonic Fibroblasts (MEFs) [34] |
Essential control for confirming Nrf2-specific effects; used to reconstitute signaling with mutant proteins. |
| PC-3 AP-1 Human Prostate Cancer Cells [32] | Model for studying AP-1 signaling and its crosstalk with Nrf2 in a cancer context. |
| Chemical Activators & Inhibitors | |
| Sulforaphane (SFN) [32] [35] | Natural isothiocyanate that modifies Keap1 cysteines (e.g., C151), leading to Nrf2 stabilization and activation. |
| tert-Butylhydroquinone (tBHQ) [34] [37] | A well-characterized Nrf2 activator used as a positive control in many experimental setups. |
| Epigallocatechin-3-gallate (EGCG) [32] | A polyphenol from green tea that can modulate both Nrf2 and AP-1 pathways. |
| MG132 [34] | Proteasome inhibitor; used to stabilize Nrf2 and accumulate ubiquitinated proteins for analysis. |
| Molecular Biology Tools | |
| ARE-Luciferase Reporter Plasmid [29] [37] | Standard tool for measuring Nrf2 transcriptional activity in high-throughput screens. |
| Keap1 Cysteine Mutant Plasmids (e.g., C151A, C273A) [35] | Used to define the functional role of specific sensor cysteines in Keap1. |
| Nrf2 Cysteine Mutant Plasmids (e.g., C235A) [34] | Used to investigate the direct redox-sensing capability of Nrf2. |
| sMaf Protein Expression Vectors [33] | Necessary partners for Nrf2 DNA binding; used to study dimerization and ARE binding. |
| Calindol Hydrochloride | Calindol Hydrochloride, CAS:729610-18-8, MF:C21H21ClN2, MW:336.9 g/mol |
| DL-Isocitric acid trisodium salt | Isocitric Acid Trisodium Salt | High Purity | RUO |
Pathway Crosstalk and Therapeutic Implications
Regulatory Interplay between Nrf2 and AP-1
Nrf2 and AP-1 do not function in isolation; they engage in extensive crosstalk. Studies using dietary chemopreventive compounds like a combination of sulforaphane and EGCG demonstrated a concerted modulation of both Nrf2 and AP-1 pathways in prostate cancer cells [32]. Bioinformatic analyses have identified conserved transcription factor binding sites in the promoter regions of Nrf2 and AP-1 components, suggesting a potential network of mutual regulation [32]. This interplay can be synergistic or antagonistic, depending on the cellular context and the specific dimeric composition of AP-1. Furthermore, the physical overlap of ARE and AP-1 sites in promoters like that of SRXN1 provides a genomic platform for this integration, allowing the cell to compute diverse stress signals into a precise transcriptional output.
Relevance to Disease and Drug Discovery
The Nrf2 pathway is a promising therapeutic target for numerous conditions driven by oxidative stress and inflammation.
- Neurodegenerative Diseases: In Alzheimer's disease (AD), Nrf2 activity is diminished, and its activation is being pursued as a strategy to combat oxidative stress and neuroinflammation. The Bach1 protein, a transcriptional repressor of Nrf2, has emerged as a complementary target, with combined Nrf2 activation and Bach1 inhibition representing a novel, multipronged therapeutic approach [38].
- Cancer: The dual role of Nrf2 in cancer presents a challenge. In healthy cells, Nrf2 activation is chemopreventive. However, cancer cells frequently harbor mutations in
KEAP1orNRF2itself, leading to constitutive Nrf2 activation that promotes tumor survival and resistance to therapy [34] [33]. This dichotomy necessitates cell-specific therapeutic strategies. - Liver and Lung Diseases: The protective role of the Nrf2-SRXN1 axis has been demonstrated in models of lung injury and various liver pathologies, including acute liver injury, alcoholic liver disease, and fibrosis [31] [29].
The development of multi-target-directed ligands (MTDLs) represents a cutting-edge approach, particularly for complex diseases like AD. For instance, chromone-containing MTDLs have been designed to simultaneously target acetylcholinesterase and activate the Nrf2/ARE pathway, demonstrating significant antioxidant effects in vitro [37].
Visualizing the Core Nrf2-Keap1-SRXN1 Signaling Pathway
The following diagram synthesizes the key components and interactions of the Nrf2-Keap1 signaling axis and its regulation of the SRXN1 gene, integrating inputs from the AP-1 pathway.
Diagram 1: Nrf2-Keap1-SRXN1 Signaling Pathway. Under basal conditions, Keap1 targets Nrf2 for proteasomal degradation. Oxidative stress modifies critical cysteine residues in Keap1, leading to Nrf2 stabilization and nuclear translocation. In the nucleus, Nrf2 dimerizes with sMaf proteins and binds the ARE to drive transcription of SRXN1. The AP-1 pathway can provide additional input at the shared ARE/AP-1 site in the SRXN1 promoter.
The transcriptional control of the antioxidant response by Nrf2 and AP-1 represents a sophisticated and layered defense system fundamental to cellular integrity. The Nrf2-Keap1 axis acts as a primary sensor for electrophiles and oxidants, launching a broad cytoprotective program. Its interplay with the context-dependent AP-1 pathway, exemplified by their co-regulation of the cysteine repair enzyme SRXN1, allows for the fine-tuning of gene expression in response to a diverse set of stimuli. Understanding the molecular nuances of this regulatory network, including the critical roles of specific cysteine residues as redox sensors, is paramount. This knowledge not only deepens our fundamental understanding of redox biology but also paves the way for rational drug design targeting Nrf2 and AP-1 in a range of debilitating diseases, from neurodegeneration to cancer. Future research, leveraging the experimental tools and models discussed, will continue to unravel the complexities of this system and its therapeutic potential.
Redox biology represents a fundamental aspect of cellular function, where reduction-oxidation (redox) reactions govern critical processes from energy production to gene expression. The term "redox" originates from the combination of "reduction" and "oxidation," describing chemical processes involving electron transfer between reactants [39]. Within biological systems, reactive oxygen species (ROS) such as superoxide (O₂•â»), hydrogen peroxide (Hâ‚‚Oâ‚‚), and hydroxyl radicals (•OH) are continuously generated through multiple sources, including the mitochondrial electron transport chain, endoplasmic reticulum, and NADPH oxidase (NOX) systems [39] [40]. For decades, ROS were predominantly viewed as toxic byproducts of metabolism that indiscriminately damage cellular macromolecules. However, our understanding has evolved to recognize that ROS also function as crucial signaling molecules that regulate a myriad of physiological processes, including insulin signaling, vascular tone, and immunometabolism [41].
This dual nature of reactive species led to the conceptual distinction between redox eustress and distress. Eustress describes oxidative stress under physiological conditions where ROS function as signaling molecules, while distress refers to the pathological state where excessive or misplaced ROS cause macromolecular damage [39]. The balance between these states is maintained by sophisticated antioxidant systems, including superoxide dismutase (SOD), catalase, glutathione peroxidase (GPx), and the thioredoxin system, which collectively maintain cellular redox homeostasis [39] [40]. The nuclear factor erythroid 2-related factor 2 (NRF2) acts as a master regulator of antioxidant responses, elevating the synthesis of key antioxidant molecules like SOD, catalase, NADPH, and glutathione (GSH) when needed [39]. This review examines the intricate mechanisms distinguishing physiological redox signaling from pathological oxidative damage, with particular emphasis on the redox regulation of protein cysteine residues and its implications for therapeutic development.
Molecular Mechanisms of Cysteine-Mediated Redox Signaling
The Cysteine Redox Switch: Post-Translational Modifications
At the heart of redox signaling lies the reversible oxidation of cysteine residues within proteins. The human proteome contains approximately 210,000 cysteine residues, with thousands exhibiting sensitivity to oxidants [40]. Cysteine residues possess a highly reactive thiol group (-SH) that undergoes a variety of reversible oxidative post-translational modifications in response to redox changes, functioning as a molecular "switch" for protein structure-function dynamics [40] [42]. The specific modifications that cysteine residues can undergo include:
- S-sulfenylation (-SOH): Formation of sulfenic acid when thiol groups are oxidized by Hâ‚‚Oâ‚‚ at low levels
- Disulfide bond formation (-S-S-): Creation of intra- or inter-molecular disulfide bonds between nearby thiols
- S-glutathionylation (-SSG): Formation of mixed disulfide bonds between sulfenic acids and glutathione
- S-persulfidation (-SSH): Modification by hydrogen sulfide (Hâ‚‚S) reacting with sulfenic acids or disulfides
- S-nitrosylation (SNO): Modification by reactive nitrogen species
- S-sulfinylation (-SOâ‚‚H): Further oxidation to sulfinic acid
- S-sulfonylation (-SO₃H): Irreversible oxidation to sulfonic acid [39] [40] [21]
Table 1: Major Reversible Cysteine Oxidative Modifications in Redox Signaling
| Modification | Chemical Structure | Regulatory Enzyme | Functional Consequence |
|---|---|---|---|
| S-sulfenylation | -SOH | --- | Protein activity modulation |
| Disulfide bond | -S-S- | Protein disulfide isomerase | Protein folding & oligomerization |
| S-glutathionylation | -SSG | Glutaredoxin | Protection from overoxidation |
| S-persulfidation | -SSH | Sulfurtransferases | Hâ‚‚S-mediated signaling |
| S-nitrosylation | -SNO | --- | NO-mediated signaling |
| S-sulfinylation | -SOâ‚‚H | Sulfiredoxin | Regulatory inactivation |
These oxidative modifications are highly dynamic and can be reversed by specific cellular reductants. For instance, sulfiredoxin-1 (SRXN1) specifically catalyzes the ATP-dependent reduction of cysteine sulfinic acid (Cys-SOâ‚‚H) in peroxiredoxins and other target proteins, playing a critical role in protecting cells from excessive oxidative damage and maintaining redox balance [21]. The reactivity of specific cysteine residues is dictated by distinct electrostatic microenvironments and subcellular localization, with solvent-exposed cysteines in certain sequence motifs being particularly susceptible to oxidation [43] [42].
Spatiotemporal Control of Redox Signaling
The biological outcome of reactive species generation depends critically on the spatiotemporal context of their production. The role of ROS is context-dependent and varies according to the cellular environment, compartmentalization, exposure period, and concentration [41]. Different cellular compartments maintain distinct redox environments and contain specialized ROS-producing and ROS-scavenging systems:
- Mitochondria: Complexes I and III of the electron transport chain are major sources of mitochondrial ROS (mtROS), alongside mitochondrial-localized proteins like NOX4, p66shc, and monoamine oxidases [41]. Mitochondria-localized SOD2 converts O₂•⻠to H₂O₂.
- Cytosol: NADPH oxidases (NOXs) are transmembrane enzymes that generate superoxide by transferring electrons from NADPH to oxygen [41]. Xanthine oxidase also produces H₂O₂ and O₂•⻠in the cytoplasm.
- Endoplasmic Reticulum: Oxidative protein folding in the ER involves ERO1 and QSOX enzymes that generate Hâ‚‚Oâ‚‚ as a byproduct when driving disulfide bond formation [40].
- Peroxisomes: House several oxidases like acyl-CoA oxidase and D-amino acid oxidase that generate Hâ‚‚Oâ‚‚ during metabolic reactions [40].
Compartmentalized ROS biosynthesis allows for localized redox regulation and ensures that redox signals are delivered to specific targets without causing widespread damage [40]. For example, communication between mitochondrial ROS and cytosolic ROS significantly affects endothelial function and angiogenesis [41]. This precise control mechanism enables specific signaling outcomes while minimizing collateral damage, representing a crucial feature distinguishing eustress from distress.
Distinguishing Redox Eustress from Distress: Molecular Determinants
Characteristics of Physiological Eustress
Redox eustress is characterized by localized, transient, and specific ROS production that activates signaling pathways essential for physiological processes. At physiological levels, ROS and hydrogen sulfide (Hâ‚‚S) act as oxidation-reduction signaling molecules that regulate a myriad of cellular processes through reversible oxidation of reactive cysteine residues in target proteins [40]. Key features of redox eustress include:
- Specific molecular targets: Redox eustress involves selective oxidation of specific cysteine residues on target proteins rather than widespread macromolecular damage. For example, quantitative cysteine redox proteomics has revealed that different cysteines within the same protein display dramatic differences in susceptibility to S-sulfenylation [43].
- Reversible modifications: Physiological signaling primarily involves reversible oxidative modifications such as S-sulfenylation, disulfide formation, and S-glutathionylation that can be rapidly reversed by cellular reduction systems [39] [40].
- Activation of adaptive responses: Eustress often activates protective transcriptional programs through transcription factors like NRF2 and FOXO, which enhance cellular antioxidant capacity and repair mechanisms [39] [40].
- Controlled production from specific enzymes: Physiological ROS generation occurs through tightly regulated systems including growth factor-activated NOX enzymes and mitochondrial respiration under normal physiological conditions [39] [41].
Notably, mild or transient increases in ROS or Hâ‚‚S levels have been shown to delay aging and extend lifespan in model organisms, demonstrating the beneficial roles of redox eustress in organismal physiology [40].
Features of Pathological Distress
In contrast to eustress, pathological redox distress is characterized by widespread, sustained, and uncontrolled oxidative damage that overwhelms cellular defense systems. The accumulation of ROS in cells directly damages biomolecules such as nucleic acids, membrane lipids, structural proteins, and enzymes, leading to cellular dysfunction or death [39]. Key aspects of redox distress include:
- Macromolecular damage: Distress causes oxidative damage to DNA (including missense mutations, truncation mutations, and DNA breakage), lipid peroxidation, and protein carbonylation [39].
- Irreversible modifications: While physiological modifications are typically reversible, distress can lead to irreversible oxidation such as sulfonylation (-SO₃H) that permanently inactivates proteins [40] [21].
- Disruption of redox-sensitive signaling: Dysregulation in redox modifications leads to aberrant redox signaling, disrupting normal cellular processes [39].
- Antioxidant system overload: Under distress, the capacity of antioxidant systems (SOD, catalase, GPx, peroxiredoxins) is exceeded, leading to loss of redox homeostasis [39] [41].
Redox distress is closely linked to the pathogenesis of a wide range of diseases, including atherosclerosis, radiation-induced lung injury, neurodegenerative diseases, cancer, and cardiovascular diseases [39] [41].
Table 2: Comparative Features of Redox Eustress and Distress
| Feature | Redox Eustress | Redox Distress |
|---|---|---|
| ROS Levels | Low, physiological | High, pathological |
| Spatial Distribution | Compartmentalized | Widespread |
| Temporal Pattern | Transient, pulsatile | Sustained, chronic |
| Molecular Targets | Specific cysteine residues | Multiple macromolecules |
| Modification Type | Reversible oxidation | Irreversible damage |
| Cellular Response | Adaptive signaling | Damage response pathways |
| Antioxidant Capacity | Sufficient | Overwhelmed |
| Biological Outcome | Physiological regulation | Cellular dysfunction |
The Redox Balance Threshold Concept
The transition between eustress and distress can be conceptualized as a threshold phenomenon where cellular antioxidant capacity is overwhelmed. The traditional view of oxidative stress as a simple imbalance between oxidants and antioxidants has evolved to recognize the sophisticated signaling functions of ROS [39]. The "Redox Code" represents a framework describing the organizing principles of redox systems, including: (1) regulation of NADH and NADPH systems in metabolism, (2) dynamic control of thiol switches in the redox proteome, (3) activation and deactivation cycles of Hâ‚‚Oâ‚‚ production, and (4) response of redox signaling to environmental changes at various cellular levels [39].
This threshold is not fixed but varies by cell type, subcellular compartment, and physiological context. For instance, immune cells like macrophages deliberately generate high ROS levels as part of their antimicrobial function, while post-mitotic cells such as neurons have lower tolerance for oxidative stress [44]. Understanding these contextual factors is essential for developing targeted therapeutic approaches that modulate redox signaling without disrupting physiological functions.
Advanced Methodologies for Mapping the Cysteine Redoxome
Chemoproteomic Workflows for S-sulfenylation Mapping
Comprehensive understanding of redox signaling requires precise mapping of cysteine oxidative modifications across the proteome. Recent advances in chemoproteomics have enabled site-specific mapping and quantification of protein S-sulfenylation at a global scale. The SulfenM workflow represents an optimized chemoproteomic approach for site-specific mapping of protein S-sulfenylation in intact cells [43]. Key steps in this methodology include:
- Selective labeling with DYn-2: Cells are treated with DYn-2 (a clickable dimedone-based probe) that selectively reacts with sulfenic acids, forming a stable covalent adduct.
- Cell lysis and protein extraction: Labeled cells are lysed, and proteins are extracted under conditions that preserve the modifications.
- Trypsin digestion: Proteins are digested with trypsin to generate peptides.
- Click chemistry conjugation: Peptides are conjugated to azido-biotin via copper-catalyzed azide-alkyne cycloaddition (CuAAC), using optimized conditions with high catalyst concentration in 30% acetonitrile in mild acid.
- Strong cation exchange (SCX) cleanup: Excess biotin reagents are removed by SCX chromatography.
- Streptavidin affinity enrichment: Biotinylated peptides are captured using streptavidin beads.
- LC-MS/MS analysis: Enriched peptides are analyzed by liquid chromatography-tandem mass spectrometry with high-energy collisional dissociation (HCD) [43].
This approach has been used to identify over 1,000 S-sulfenylation sites on more than 700 proteins in intact cells, providing unprecedented coverage of the S-sulfenylome [43]. The method benefits from diagnostic fragment ions (m/z 368.16, 366.15, 336.19) that increase confidence in S-sulfenylated peptide identifications [43].
Diagram 1: SulfenM chemoproteomic workflow for mapping protein S-sulfenylation.
Quantitative S-sulfenylation Analysis (SulfenQ)
For quantitative assessment of dynamic changes in S-sulfenylation, the SulfenQ methodology has been developed. This approach enables comparative analysis of S-sulfenylation patterns under different experimental conditions [43]:
- Metabolic labeling: Cells in two different conditions are labeled with light and heavy DYn-2 isotopomers, respectively.
- Sample combination: Cells are combined after labeling and digested with trypsin.
- Biotin tagging and enrichment: Tagged peptides are biotinylated, captured, and enriched as in the SulfenM workflow.
- Quantitative MS analysis: Relative changes in protein S-sulfenylation are determined by the ratio of extracted ion current peak intensities for heavy and light isotope-labeled DYn-2-tagged peptides using MS1 filtering.
This quantitative approach has revealed that different cysteine residues within the same protein can display dramatically different responses to oxidative stimuli. For example, in fatty acid synthase (FASN), nine cysteines showed S-sulfenylation ratios distributed over a >60-fold range in response to Hâ‚‚Oâ‚‚ treatment [43]. Similarly, in PRDX6, C91 showed a dramatic increase in S-sulfenylation after Hâ‚‚Oâ‚‚ treatment (RH/L = 9.15), while the active site C47 showed decreased S-sulfenylation (RH/L = 0.07) due to susceptibility to overoxidation [43].
Cysteine Redoxome Mapping in Disease Models
Application of these methodologies in disease-relevant contexts has provided insights into pathological redox alterations. For example, a comprehensive cysteine redoxome mapping study in livers of male mice fed a high-fat/high-sucrose diet (HFHSD) pinpointed over 5,000 oxidized and reduced cysteine residues [42]. While the global distribution of oxidized and reduced cysteine residues remained stable across both diet groups, 169 specific cysteine residues exhibited dynamic redox changes in response to HFHSD, mapping to 35 KEGG pathways central to redox balance and energy homeostasis [42]. Structural analyses demonstrated that cysteine residues sensitive to HFHSD-induced oxidation were enriched in mitochondria and cytosol, while those sensitive to HFHSD-induced reduction were found in extracellular regions and participated in disulfide bond formation [42].
Table 3: Essential Research Reagents for Cysteine Redoxome Studies
| Reagent / Tool | Specific Example | Application / Function |
|---|---|---|
| Sulfenic Acid Probe | DYn-2 | Selective labeling of S-sulfenylated cysteines |
| Isotope-labeled Probe | Light/Heavy DYn-2 | Quantitative comparison of S-sulfenylation |
| Click Chemistry Reagents | Azido-biotin, TBTA, CuSOâ‚„ | Biotin tagging for affinity enrichment |
| Affinity Matrix | Streptavidin beads | Enrichment of biotinylated peptides |
| Separation Media | Strong cation exchange | Cleanup of tagged peptides |
| Protease | Trypsin | Protein digestion for MS analysis |
| MS Instrumentation | Q-Exactive with HCD | High-accuracy identification and quantification |
Redox Regulation of Specific Biological Processes: From Physiology to Pathology
Cell Cycle Regulation
Redox signaling plays a critical role in coordinating cell proliferation and fate decisions. Cell-cycle-resolved S-sulfenylation proteomics has revealed that although overall ROS levels rise during cell cycle progression, dynamic S-sulfenylation is restricted to a subset of cysteines [45]. Among these, a critical redox-sensitive cysteine residue (C41) in the cyclin-dependent kinase (CDK) inhibitor p21 serves as a redox switch that regulates the interaction of p21 with CDK2 and CDK4, controlling a double-negative feedback loop that determines p21 stability [45]. When C41 remains reduced, p21's half-life increases in the G2 phase, resulting in more p21 inheritance to daughter cells, suppressing proliferation and promoting senescence after irradiation [45]. This mechanism demonstrates how specific cysteine oxidation events can influence cell fate decisions in response to environmental stimuli.
Genomic Stability
Redox signaling significantly impacts genomic stability through both direct DNA damage and regulation of DNA repair proteins. Oxidative stress promotes the accumulation of ROS that can chemically induce DNA missense mutations, truncation mutations, or DNA breakage during replication or transcription [39]. Beyond direct damage, redox signaling finely regulates the repair of DNA damage through redox modifications of DNA repair-related proteins [39]. For instance:
- Double-strand break repair: The activation of ataxia-telangiectasia mutated protein kinase (ATM) is triggered by the Mre11-Rad50-Nbs1 (MRN) complex when damage occurs, facilitating repair of double-strand breaks. The C-terminal kinase domain of ATM undergoes autophosphorylation in a redox-regulated manner [39].
- Base excision repair: Multiple enzymes in base excision repair pathways contain redox-sensitive cysteine residues that modulate their activity in response to oxidative stress [39].
These findings establish redox signaling as a crucial regulator of genomic integrity, with implications for cancer, aging, and degenerative diseases.
Innate Immunity and Inflammation
Redox signaling plays a central role in regulating macrophage and neutrophil function, integrating ROS, reactive nitrogen species (RNS), and reactive sulfur species (RSS) to modulate innate immune responses [44]. Reactive species modulate diverse cellular processes in phagocytes, including differentiation, metabolic adaptation, cytokine production, and cell death [44]. Key redox-regulated processes in immunity include:
- Inflammasome activation: ROS production triggers NLRP3 inflammasome assembly, promoting cytokine secretion
- Metabolic reprogramming: Redox signals regulate the shift between oxidative phosphorylation and glycolysis in immune cells
- Phagocytosis and microbial killing: NADPH oxidase-derived ROS directly contribute to pathogen elimination
- Resolution of inflammation: Redox changes facilitate the transition from pro-inflammatory to pro-resolving phenotypes [41] [44]
Antioxidant systems, including the glutathione and thioredoxin systems, play essential roles in maintaining redox balance, counteracting excessive oxidants, and preserving immune cell function [44]. Dysregulation of these redox control mechanisms contributes to chronic inflammatory diseases and impaired host defense.
Diagram 2: Redox signaling pathway from stimulus to cellular response.
Therapeutic Implications and Future Directions
Challenges in Antioxidant Therapy
The conceptual distinction between redox eustress and distress has profound implications for therapeutic development. Traditional antioxidant approaches using broad-spectrum antioxidants like vitamin E have largely failed in clinical trials, likely because they non-specifically scavenge both physiological signaling ROS and pathological damaging ROS [39] [41]. These approaches lack specificity, inability to target the main sources of ROS, and tendency to overlook the physiological roles of ROS in signaling and defense [41]. A more nuanced approach to antioxidant interventions is needed—one that supports essential physiological redox processes yet affords protection against the onset and development of diseases [41].
Targeted Redox Therapeutics
Emerging strategies aim to develop targeted interventions that specifically modulate pathological redox signaling without disrupting physiological eustress:
- Small molecule inhibitors: Emerging small molecule inhibitors that target specific cysteine residues in redox-sensitive proteins have demonstrated promising preclinical outcomes, setting the stage for forthcoming clinical trials [39].
- SRXN1-targeted therapies: Sulfiredoxin-1 represents an attractive therapeutic target due to its central role in regulating oxidative stress and its involvement in multiple disease processes [21]. Modulation of SRXN1 activity may offer a novel therapeutic avenue to mitigate cysteine oxidation and improve clinical outcomes in various disease contexts [21].
- NRF2 activators: Pharmacological activation of the NRF2 pathway enhances expression of multiple antioxidant genes, including SRXN1, providing a coordinated cytoprotective response [39] [21].
- Isoform-specific NOX inhibitors: Development of compounds that specifically inhibit pathological NOX isoforms without affecting physiological ROS signaling [41].
These targeted approaches require a deep, context-specific understanding of redox signaling, particularly the roles of redox-sensitive proteins and their specific modification sites [39].
Future Perspectives in Redox Medicine
Future advances in redox medicine will depend on several key developments:
- Spatiotemporally precise redox probes: Continued development of advanced chemoproteomic methods with improved spatial and temporal resolution will enable more precise mapping of redox dynamics in living systems.
- Single-cell redoxomics: Technologies for assessing redox states at the single-cell level will reveal cell-to-cell heterogeneity in redox responses.
- Redox epigenetics: Further exploration of how redox signaling influences epigenetic modifications will uncover novel regulatory mechanisms [39].
- Context-specific therapeutic design: Treatments tailored to specific pathological contexts where redox imbalance plays a primary role, such as radiation-induced lung injury and paraquat poisoning [39].
As our understanding of the intricate relationship between oxidative stress and disease pathogenesis deepens, we can increasingly leverage these insights to optimize therapeutic strategies in clinical practice [39]. The growing catalog of redox-sensitive cysteines and their specific roles in health and disease provides a rich resource for drug discovery and therapeutic target identification.
The distinction between redox eustress and distress represents a fundamental paradigm in understanding how oxidative processes influence health and disease. Physiological redox eustress, mediated through reversible cysteine oxidation, regulates essential cellular processes including proliferation, metabolism, and adaptive responses. In contrast, pathological redox distress involves widespread oxidative damage that disrupts cellular function and contributes to disease pathogenesis. The precise molecular context—including specific cysteine residues affected, spatial localization of ROS production, temporal dynamics of oxidation, and cellular antioxidant capacity—determines whether redox processes yield adaptive signaling or detrimental damage.
Advanced chemoproteomic methods now enable comprehensive mapping of the cysteine redoxome, revealing thousands of dynamically regulated cysteine residues that respond to physiological and pathological stimuli. These technologies provide unprecedented insights into the molecular mechanisms of redox regulation and identify potential therapeutic targets for diseases characterized by redox imbalance. Future therapeutic development must move beyond non-specific antioxidant approaches toward targeted strategies that selectively modulate pathological redox signaling while preserving beneficial redox regulation. Through continued elucidation of the complex networks governing redox homeostasis, we can develop more effective interventions for the wide range of diseases associated with oxidative stress.
Mapping the Redoxome: Advanced Proteomic and Computational Tools
Redox signaling is a fundamental mediator of adaptive responses in biological systems, influencing metabolic regulation, stress responses, and developmental processes through thiol-based oxidative post-translational modifications (oxiPTMs) of redox-sensitive proteins [46] [47]. These modifications, particularly those involving cysteine (Cys) residues, function as molecular switches that dynamically alter protein function, structure, and interactions [46]. The central challenge in studying these modifications lies in their dynamic and reversible nature, often occurring at low stoichiometry within complex biological samples [48]. Mass spectrometry-based redox proteomics has emerged as a powerful technological platform that enables precise detection, quantification, and functional annotation of these oxidative modifications in physiological contexts [46] [49]. This technical guide comprehensively details three cornerstone methodologies—Isotope-Coded Affinity Tags (ICAT), Resin-Assisted Capture (RAC), and the Biotin-Switch Assay—that have revolutionized our ability to interrogate the cysteine redoxome within the broader context of redox regulation research.
Core Methodologies and Experimental Protocols
Isotope-Coded Affinity Tags (ICAT)
Principle and Mechanism: The ICAT methodology utilizes tags with identical structure but different isotopic compositions (e.g., light ([¹²C]) and heavy ([¹³C]) to differentially label reduced cysteine thiols from two distinct biological samples [46] [50]. This approach allows for precise quantification of oxidation states by comparing the abundance of peptides from oxidized versus reduced samples.
Experimental Workflow:
- Sample Preparation: Proteins are extracted under denaturing conditions to preserve native redox states.
- Blocking of Oxidized Cysteines: Free thiols in the control sample are blocked with a conventional alkylating agent.
- Reduction of Oxidized Cysteines: reversibly oxidized cysteine residues in the experimental sample are reduced using agents such as tris(2-carboxyethyl)phosphine (TCEP) or dithiothreitol (DTT).
- ICAT Labeling: The newly reduced thiols in the experimental sample are labeled with heavy ICAT reagents, while the control sample is labeled with light ICAT reagents.
- Sample Combination and Proteolysis: The differentially labeled samples are combined in a 1:1 ratio and digested into peptides.
- Affinity Enrichment: Cysteine-containing peptides are isolated using the affinity tag (e.g., biotin-avidin chemistry).
- LC-MS/MS Analysis: Enriched peptides are analyzed by liquid chromatography-tandem mass spectrometry, with quantification achieved by comparing the relative abundances of light and heavy peptide pairs [46] [50].
Resin-Assisted Capture (RAC)
Principle and Mechanism: RAC employs thiol-reactive resins (typically thiopropyl sepharose) to directly capture reduced cysteine residues following the specific reduction of oxidized cysteines [46] [51]. This method enables selective enrichment of previously oxidized proteins or peptides without requiring specific chemical tags.
Experimental Workflow:
- Free Thiol Blocking: Free cysteine thiols are blocked with alkylating agents such as N-ethylmaleimide (NEM) or iodoacetamide (IAM) under denaturing conditions.
- Reduction of Reversible OxiPTMs: Reversibly oxidized cysteine residues (e.g., disulfides, S-nitrosylation) are selectively reduced using appropriate reducing agents.
- Resin Capture: Newly exposed thiols are captured onto thiol-reactive resin through disulfide bond formation.
- Stringent Washing: Non-specifically bound proteins are removed through extensive washing.
- Elution: Captured proteins/peptides are released from the resin using reducing agents such as DTT or β-mercaptoethanol.
- Proteomic Analysis: Eluted proteins are digested (if captured at protein level) and analyzed by LC-MS/MS [46] [51].
Biotin-Switch Assay (BSA)
Principle and Mechanism: The Biotin-Switch Assay is particularly effective for detecting specific oxidative modifications, most notably S-nitrosylation [46]. This three-step method converts labile oxidative modifications to a stable biotin tag, enabling enrichment and detection.
Experimental Workflow:
- Thiol Blocking: Free thiols are blocked with methyl methanethiosulfonate (MMTS) or N-ethylmaleimide (NEM) under dark conditions to prevent light-induced decomposition of S-nitrosothiols.
- Selective Reduction: The specific oxidative modification of interest (e.g., S-nitrosothiols) is selectively reduced using ascorbate or similar selective reductants.
- Biotin Labeling: Newly exposed thiols are labeled with biotin-containing thiol-reactive reagents such as N-[6-(biotinamido)hexyl]-3'-(2'-pyridyldithio) propionamide (biotin-HPDP).
- Affinity Purification: Biotin-labeled proteins are captured using streptavidin or neutravidin resins.
- Elution and Analysis: Proteins are eluted with reducing agents and analyzed by immunoblotting or mass spectrometry [46] [52].
Comparative Analysis of Techniques
Table 1: Comprehensive Comparison of Redox Proteomics Methods
| Parameter | ICAT | RAC | Biotin-Switch Assay |
|---|---|---|---|
| Primary Applications | Quantitative profiling of general cysteine oxidation states [46] | Enrichment of reversibly oxidized cysteine residues [46] [51] | Specific detection of S-nitrosylation and other selective oxiPTMs [46] |
| Quantification Capability | Excellent (isotopic labeling) [50] | Semi-quantitative (label-free) | Semi-quantitative (can be combined with isotopic tags) |
| Sensitivity | High (due to enrichment) | Moderate to High | High (due to high-affinity biotin-streptavidin interaction) |
| Specificity | Broad (all reduced cysteines) | Broad (reversibly oxidized cysteines) | Targeted (specific to selected oxiPTMs) |
| Key Advantages | Direct quantitative comparison; Reduced complexity after enrichment | Versatile for various oxiPTMs; No special reagents required | Highly specific for target modifications; Compatible with multiple detection methods |
| Limitations | Requires specific reagents; Higher cost | Limited quantification precision; Potential for non-specific binding | Multi-step process risk of artifacts; Incomplete blocking causes false positives |
| Compatible OxiPTMs | General redox state assessment | S-glutathionylation, disulfides, sulfenic acids [46] [48] | S-nitrosylation, sulfenic acids (with specific reduction) [46] |
Table 2: Research Reagent Solutions for Redox Proteomics
| Reagent Category | Specific Examples | Function and Application |
|---|---|---|
| Thiol Blocking Agents | N-ethylmaleimide (NEM), Iodoacetamide (IAM), Methyl methanethiosulfonate (MMTS) [48] [51] | Alkylates free thiols to prevent further oxidation during sample processing; critical first step in all three methods |
| Reducing Agents | Tris(2-carboxyethyl)phosphine (TCEP), Dithiothreitol (DTT), β-mercaptoethanol [51] | Reduces reversible oxidative modifications to free thiols for subsequent labeling |
| Selective Reductants | Ascorbate, Arsenite [46] | Specifically reduces particular oxiPTMs (e.g., ascorbate for S-nitrosylation) |
| Affinity Tags | ICAT reagents (biotin-based), HPDP-biotin, IAM-alkyne probes [48] [51] | Provides handle for enrichment of labeled peptides/proteins |
| Enrichment Matrices | Streptavidin/NeutrAvidin beads, Thiopropyl sepharose resin [46] [51] | Solid supports for selective capture of tagged proteins/peptides |
| Isotopic Labels | ¹²C/¹³C ICAT tags, Dimethyl labeling [50] | Enables quantitative comparison between samples by mass spectrometry |
Technical Considerations and Applications in Research
Critical Methodological Considerations
Optimization of Cysteine Blocking: Efficient and complete blocking of free thiols is paramount for all three methods. Inadequate blocking leads to significant false-positive results [51]. The alkylation reaction requires careful optimization of pH (typically pH 7.4) to balance reaction efficiency with preservation of thioester-linked modifications [51]. N-ethylmaleimide (NEM) has been widely adopted due to its high reactivity with cysteine thiols, with some protocols implementing sequential rounds of NEM treatment to ensure complete blocking [51].
Specificity in Reduction Steps: The Biotin-Switch Assay's specificity for S-nitrosylation depends critically on the selective reduction step. Ascorbate effectively reduces S-nitrosothiols with minimal effect on other oxidative modifications [46]. However, ascorbate concentration and reaction time must be optimized to prevent non-specific reduction.
Quantification Strategies: While ICAT provides built-in quantification through isotopic labeling, RAC and basic Biotin-Switch protocols are primarily qualitative. For quantitative applications, these methods can be combined with isobaric tags (e.g., TMT, iTRAQ) or label-free approaches, though these require careful normalization and experimental design [46] [50].
Applications in Biological Research
These techniques have enabled significant advances across diverse research domains. In plant biology, redox proteomics has elucidated mechanisms underlying development, seed germination, fruit ripening, and stress adaptation [46]. For example, Wang et al. utilized iodoTMT (a related methodology) to identify 70 redox-sensitive peptides during tomato fruit ripening, linking oxidation of specific enzymes like polygalacturonase 2A to fruit softening [46].
In neurodegenerative disease research, redox proteomics has identified oxidatively modified proteins in Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis [49] [52]. The accumulation of oxidatively modified proteins has been implicated in the pathogenesis of these conditions, with specific modifications like S-nitrosylation of peroxiredoxin 2 observed in Parkinson's disease brains [52].
In cancer biology, redox proteomics has revealed how cancer cells exploit redox signaling for proliferation and survival. The SICyLIA method (a related workflow) has been applied to models of fumarate hydratase deficiency, revealing how chronic oxidative stress induces specific metabolic adaptations through oxidation of distinct metabolic proteins [50].
ICAT, RAC, and the Biotin-Switch Assay represent foundational methodologies that have propelled redox proteomics into a central role in biological research. Each technique offers distinct advantages and limitations, making them complementary rather than competitive. ICAT excels in quantitative applications, RAC provides versatile enrichment of various oxiPTMs, and the Biotin-Switch Assay offers specificity for targeted modifications. As mass spectrometry technology continues to advance, with improvements in sensitivity, speed, and data analysis algorithms, these methodologies will undoubtedly evolve, providing increasingly comprehensive insights into the redox regulation of biological systems. For drug development professionals, these techniques offer powerful tools for identifying novel therapeutic targets and assessing target engagement of redox-modulating compounds.
Redox signaling, mediated through the reversible oxidation of protein cysteine residues, is a fundamental regulatory mechanism in cellular biology [46] [53]. These oxidative post-translational modifications (oxiPTMs), including S-sulfenylation, S-nitrosation, and S-glutathionylation, act as molecular switches that dynamically alter protein function, structure, and interactions in response to metabolic changes and environmental stresses [46] [54]. However, the analysis of these modifications presents significant technical challenges due to their low abundance, transient nature, and dynamic reversibility, often making them difficult to detect with conventional biochemical methods [46] [53].
The development of advanced mass spectrometry-based redox proteomics has revolutionized our ability to capture and quantify these elusive modifications [46]. Among the most powerful approaches are OxICAT (Isotope-Coded Affinity Tag) and iodoTMT (Isobaric Tandem Mass Tag) techniques, which enable highly precise, site-specific quantification of cysteine oxidation states within complex proteomes [46] [55]. These methods have become indispensable tools for moving from merely detecting oxiPTMs to understanding their functional significance in redox signaling networks, stress adaptation, and disease progression [46] [56]. This whitepaper provides a comprehensive technical guide to these foundational techniques, detailing their methodologies, applications, and implementation considerations for the research community.
Core Principles of Site-Specific Redox Quantification
OxICAT: In Vivo Oxidation Status Quantification
The OxICAT method employs a sophisticated differential alkylation strategy with isotopically labeled tags to determine the in vivo oxidation status of cysteine thiols [56]. Its key differentiator is the preservation of the native redox state through immediate acid trapping of cellular material, which prevents artifactual oxidation during sample preparation [56] [57]. The multi-step procedure involves: (1) blocking reduced thiols in denatured protein extracts with the light ICAT reagent (containing ( ^{12}C )); (2) reducing previously oxidized thiols with chemical reductants like Tris(2-carboxyethyl)phosphine (TCEP); and (3) labeling the newly reduced thiols with the heavy ICAT reagent (containing ( ^{13}C )) [56]. After tryptic digestion, affinity purification of ICAT-labeled peptides enables liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, where the relative quantification of light and heavy peptide pairs directly reveals the oxidation percentage at specific cysteine sites [56].
iodoTMT: Multiplexed Comparative Analysis
The iodoTMT platform utilizes isobaric tags to enable multiplexed comparison of cysteine oxidation across multiple samples simultaneously [46] [55]. In this approach, reduced cysteine thiols are first blocked with iodoacetamide-based TMT reagents, which have the same total mass but yield distinct reporter ions upon fragmentation during MS/MS analysis [46]. After reducing reversibly oxidized thiols, the newly accessible thiols are labeled with a different set of TMT reagents [46] [55]. This design allows researchers to pool samples early in the workflow, reducing technical variability and enabling higher throughput [46]. The relative intensities of the reporter ions in MS/MS spectra provide quantitative information about oxidation states across multiple conditions in a single experiment, making iodoTMT particularly valuable for time-course studies or complex experimental designs [46] [55].
Experimental Protocols and Workflow Implementation
Step-by-Step OxICAT Methodology
The OxICAT protocol requires meticulous execution to preserve the native redox state of cysteines [56] [57]:
Cell Culture and Treatment: Grow yeast cells to mid-logarithmic phase in appropriate media. For oxidative stress treatment, apply 1 mM H(2)O(2) for 30 minutes to induce reversible oxidation without significant cell death [56].
Rapid Metabolic Quenching and Protein Precipitation: Immediately quench cellular metabolism by adding trichloroacetic acid (TCA) to a final concentration of 10-20% (w/v). Incubate samples on ice for at least 20 minutes to precipitate proteins while preserving in vivo oxidation states [56] [57].
Differential Alkylation with ICAT Reagents:
- Step 1 - Label Reduced Thiols: Resuspend protein pellets in denaturing buffer (8 M urea, 100 mM N-ethylmaleimide [NEM], 10 mM EDTA, 2% SDS) with light ICAT reagent. Incubate at 50°C for 30 minutes with shaking at 1000 rpm [56] [57].
- Step 2 - Reduce Oxidized Thiols: Precipitate proteins again with TCA to remove excess light ICAT. Reduce reversibly oxidized disulfides and other modifications by resuspending in TUNES buffer without NEM but with 5.5 mM TCEP. Incubate at 50°C for 30 minutes [56].
- Step 3 - Label Newly Accessible Thiols: Add heavy ICAT reagent to the reduced samples and incubate at 50°C for 30 minutes at 1200 rpm [56].
Protein Digestion and Affinity Purification: Digest proteins with sequencing-grade trypsin overnight at 37°C. Isolate ICAT-labeled peptides using avidin affinity chromatography, which specifically captures the biotin moiety present in ICAT reagents [56] [57].
LC-MS/MS Analysis and Data Processing: Analyze peptides using nanoflow LC-MS/MS. Quantify the relative abundance of light and heavy peptide pairs using computational platforms like MaxQuant, and calculate oxidation percentages using the formula: % Oxidation = (Heavy/(Heavy + Light)) × 100 [56].
Comprehensive iodoTMT Workflow
The iodoTMT protocol enables multiplexed analysis of up to 16 samples [46] [55]:
Sample Preparation and Reduction: Lyse cells or tissues in appropriate buffers. Determine protein concentration and aliquot equal amounts for each experimental condition [55].
Blocking Reduced Cysteines: Add iodoacetyl TMTpro reagents to block free thiols. Incubate in the dark at room temperature for 2 hours with gentle agitation [46] [55].
Reduction of Oxidized Cysteines: Reduce reversibly oxidized cysteines by adding DTT to a final concentration of 5-10 mM. Incubate at 37°C for 30-60 minutes [55].
Labeling Newly Reduced Cysteines: Add a different set of iodoacetyl TMTpro tags to label the newly accessible thiols. Incubate in the dark for 2 hours at room temperature [46] [55].
Sample Pooling and Digestion: Combine all TMT-labeled samples in equal ratios. Digest the pooled sample with trypsin overnight at 37°C [46].
High-pH Fractionation and LC-MS/MS: Fractionate peptides using high-pH reverse-phase chromatography to reduce complexity. Analyze fractions by LC-MS/MS on an Orbitrap mass spectrometer. Quantify reporter ion intensities in MS/MS spectra for relative quantification of oxidation states across samples [46] [55].
Table 1: Key Research Reagent Solutions for Redox Proteomics
| Reagent/Tool | Function | Application Context |
|---|---|---|
| ICAT Reagents (light ( ^{12}C ) / heavy ( ^{13}C )) | Differential alkylation for quantifying reduced vs. oxidized thiol ratios | OxICAT method; identifies site-specific oxidation percentages [56] |
| Iodoacetyl TMTpro Tags | Isobaric tags for multiplexed quantification of cysteine oxidation | iodoTMT platform; enables comparison of up to 16 samples [46] [55] |
| Trichloroacetic Acid (TCA) | Acid trapping for preserving in vivo oxidation states during sample preparation | Essential first step in both OxICAT and iodoTMT workflows [56] [57] |
| Tris(2-carboxyethyl)phosphine (TCEP) | Reducing agent for reversing disulfide bonds and other reversible oxiPTMs | Critical step in differential alkylation to reveal oxidized thiols [56] [57] |
| Sequencing-Grade Trypsin | Proteolytic enzyme for digesting proteins into peptides for MS analysis | Standard protein digestion in both protocols [56] [57] |
Research Applications and Key Findings
Quantitative redox proteomics has yielded transformative insights across diverse biological systems. The tables below summarize exemplary applications of OxICAT and iodoTMS technologies.
Table 2: Key Research Findings Using OxICAT and iodoTMT Technologies
| Biological System | Technique | Key Finding | Biological Impact | Reference |
|---|---|---|---|---|
| Saccharomyces cerevisiae (yeast) | OxICAT | Identified redox switches in translation machinery; mapped >4,300 unique cysteine residues in >2,200 proteins | Mitochondrial ROS reversibly control global protein synthesis [56] | |
| Tomato fruit | iodoTMT | Identified 70 redox-sensitive peptides from 51 proteins during ripening; oxidation of PG2A and E8 enzymes | Direct molecular link between redox modification and fruit softening [46] [53] | |
| Poplar seedlings | iodoTMT | Discovered 118 and 101 differentially modified cysteine sites under pHBA and H(2)O(2) stress, respectively | Oxidative modifications regulate MAPK signaling and metabolic adaptation [55] | |
| Arabidopsis guard cells | TMT-based redox proteomics | Redox-sensitive proteins involved in photosynthesis and lipid binding respond to bacterial elicitor flg22 | Redox-dependent regulation of plant immune responses [46] |
Technical Considerations and Comparative Analysis
OxICAT vs. iodoTMT: Strategic Selection
Choosing between OxICAT and iodoTMT requires careful consideration of research goals and technical constraints:
OxICAT Advantages: Provides absolute quantification of oxidation percentages, superior for determining the intrinsic redox potential of specific cysteine residues. The affinity enrichment step enhances sensitivity for detecting low-abundance cysteine-containing peptides [56].
iodoTMT Advantages: Enables multiplexing (currently up to 16-plex with TMTpro), reducing analytical time and inter-sample variability. Higher throughput makes it ideal for screening multiple conditions, time courses, or biological replicates [46] [55].
Limitations: OxICAT's requirement for avidin affinity purification adds complexity and potential for sample loss. iodoTMT's multiplexing, while efficient, is limited by the number of available TMT channels and can suffer from ratio compression due to co-isolation of interfering peptides [46] [56].
Complementary and Emerging Methods
Other significant techniques enrich the redox proteomics toolkit:
SICyLIA (Stable Isotope Cysteine Labeling with Iodoacetamide): A proteome-wide approach without enrichment steps, using light/heavy iodacetamide (IAM) for unbiased quantification. Successfully identified oxidation in metabolic proteins under acute and chronic oxidative stress [58].
OxSWATH: Combines differential alkylation with SWATH-MS for label-free quantification, simultaneously analyzing redox changes and total protein levels, providing integrated datasets [59].
Targeted Approaches (OxMRM): Using multiple reaction monitoring for highly sensitive, specific quantification of predefined cysteine residues in low-abundance proteins, ideal for validating discoveries from global screens [57].
Visualization of Experimental Workflows
OxICAT and iodoTMT represent cornerstone methodologies in the quantitative analysis of cysteine redox modifications, each offering distinct advantages for specific research applications. OxICAT excels in determining precise oxidation percentages of cysteine residues under physiological conditions, while iodoTMT provides powerful multiplexing capabilities for comparative studies across multiple experimental conditions [46] [56]. The integration of these techniques with emerging computational approaches, including machine learning algorithms like CysQuant and DLF-Sul, is further expanding our predictive capabilities for identifying redox-sensitive residues and modeling redox signaling networks [46] [53].
As redox proteomics continues to evolve, several promising directions are emerging. The development of more sensitive mass spectrometers will enhance detection limits for low-abundance redox modifications. New chemical biology tools, such as photocaged cysteine sulfoxides, enable precise, site-specific manipulation of redox events to establish causal relationships [6]. Additionally, the integration of redox proteomics with other omics technologies (transcriptomics, metabolomics) through systems biology approaches will provide increasingly holistic views of redox regulatory networks [46] [53]. These advances, coupled with the foundational techniques detailed in this whitepaper, promise to accelerate both our basic understanding of redox biology and the translation of this knowledge to therapeutic interventions for oxidative stress-related pathologies and precision agriculture applications.
The study of redox regulation, which centers on the reversible oxidation of protein cysteine residues, is undergoing a profound transformation driven by artificial intelligence (AI) and machine learning (ML). Redox signaling operates through rapid and reversible oxidation of reactive cysteine thiols, which function as molecular switches to control protein function, localization, and interaction in response to oxidative stimuli [40]. For decades, the field has been challenged by the transient nature of these modifications and the difficulty of capturing their dynamics in living systems. The integration of computational models with advanced mass spectrometry-based proteomics is now bridging this gap, enabling researchers to move from descriptive biology to predictive and manipulative science [54] [53]. This whitepaper examines two pivotal technologies—CysQuant for experimental quantification and DLF-Sul for computational prediction—that exemplify this transformation and their applications in basic research and drug development.
Within the broader thesis of cysteine redox regulation research, these tools represent complementary approaches: CysQuant provides high-fidelity experimental measurement of cysteine oxidation states, while DLF-Sul offers predictive power for identifying novel redox-sensitive sites across proteomes. Together, they form a synergistic toolkit for comprehensively mapping and validating the cysteine redoxome—the complete set of redox-modified cysteine residues within a biological system. For research and drug development professionals, understanding the capabilities, applications, and integration of these technologies is crucial for advancing both fundamental knowledge and therapeutic strategies targeting redox dysregulation in disease.
Computational Prediction Tools for Redox-Dependent PTMs
Computational tools have become essential for predicting redox-dependent post-translational modifications (oxiPTMs), particularly when experimental validation is limited or resource-intensive [53]. These tools leverage machine learning algorithms trained on experimentally verified modification sites to identify patterns and features associated with redox-sensitive cysteines. The emergence of deep learning architectures has significantly enhanced prediction accuracy by enabling models to automatically learn relevant features from protein sequence and structural data without manual feature engineering.
The predictive modeling ecosystem for cysteine modifications has expanded substantially, with tools now available for various specific modifications including S-nitrosylation, S-sulfenylation, S-glutathionylation, persulfidation, and disulfide bond formation [54] [53]. These computational approaches are transforming redox biology from a largely descriptive field into one that can predict and manipulate redox-dependent processes, offering exciting possibilities for understanding disease mechanisms and developing targeted interventions [53].
Table 1: Key Computational Tools for Predicting Cysteine Redox Modifications
| Tool Name | Prediction Focus | Underlying Algorithm | Key Features | Applications in Research |
|---|---|---|---|---|
| DLF-Sul [53] | S-sulfenylation sites | Machine Learning/Deep Learning | High-precision prediction of sulfenic acid formation | Identifies redox-sensitive signaling nodes; prioritizes experimental validation |
| CysQuant [60] [61] | Cysteine oxidation degree & protein abundance | Mass spectrometry with computational analysis | Simultaneous quantification of oxidation states and protein levels | Validation of redox proteomics; study of redox dynamics in stress responses |
| pCysMod [62] | Multiple cysteine modifications | Deep Learning with Particle Swarm Optimization | Predicts S-nitrosylation, S-palmitoylation, S-sulfenylation, S-sulfhydration, S-sulfinylation | Comprehensive analysis of modification crosstalk; systems biology approaches |
| BiGRUD-SA [53] | Sulfenylation prediction | Machine Learning | Specialized for plant redox proteomics | Crop improvement research; plant stress physiology studies |
These computational tools share a common workflow beginning with feature extraction from protein sequences, including position-specific scoring matrices, amino acid composition, and structural attributes. The models are then trained on curated datasets of experimentally verified modification sites, with performance validation through cross-validation and independent testing [62]. The predictive performance of these models has reached impressive levels, with tools like pCysMod achieving AUC values of 0.793-0.876 for various cysteine modifications in cross-validation studies [62].
CysQuant: Experimental Quantification of Cysteine Oxidation
Methodology and Technical Workflow
CysQuant represents a methodological breakthrough in experimental redox proteomics, enabling simultaneous quantification of cysteine oxidation degrees and protein abundances using data-dependent (DDA) or data-independent acquisition (DIA) mass spectrometry [60]. The core innovation lies in its differential labeling approach, which utilizes light and heavy iodoacetamide isotopologues to distinguish between reduced and reversibly oxidized cysteine residues within complex biological samples [60] [61].
The technical workflow begins with protein extraction under controlled conditions that preserve the native redox state. Cysteine residues are then subjected to differential labeling: reduced thiols are blocked with light iodoacetamide, while reversibly oxidized cysteines (including disulfides) are first reduced followed by labeling with heavy iodoacetamide. This differential tagging creates a mass signature that allows the mass spectrometer to distinguish and quantify the reduced and oxidized populations of each cysteine-containing peptide [60]. When combined with isobaric labeling for protein quantification, this approach enables researchers to determine both the oxidation percentage at specific cysteine sites and the absolute abundance of the corresponding proteins—two critical parameters for understanding redox regulation in physiological and pathological contexts.
Table 2: Key Research Reagents for CysQuant Implementation
| Reagent / Material | Function in Experimental Workflow | Technical Specifications | Application Context |
|---|---|---|---|
| Light/Heavy Iodoacetamide Isotopologues [60] | Differential labeling of reduced vs. oxidized cysteines | Chemical purity >95%; Defined mass difference (e.g., +3 Da for duplex) | Fundamental to CysQuant workflow; enables MS discrimination |
| Reducing Agents (TCEP/DTT) | Reduction of reversible oxidations for heavy labeling | Freshly prepared; Concentration optimized to avoid over-reduction | Specific reduction of reversible oxiPTMs (disulfides, sulfenic acids) |
| IodoTMT / TMT Reagents [53] | Multiplexed protein quantification | 6-16 plex kits; Manufacturer's protocol | Enables multiplexed experimental designs and protein abundance measurement |
| Predicted Spectral Libraries [60] | Peptide identification in DIA analysis | Generated in silico; Organism-specific | Critical for DIA-MS data analysis; enables identification without experimental libraries |
| Solid-phase Extraction Cartridges | Sample cleanup and desalting | C18 material; Compatible with sample volume | Post-labeling cleanup before MS analysis |
Applications and Experimental Validation
In practice, CysQuant has demonstrated remarkable utility in characterizing redox dynamics in complex biological systems. Application to Arabidopsis thaliana revealed an average of 18% cysteine oxidation across the proteome, with a subset of highly oxidized cysteines participating in disulfide bridges as predicted by AlphaFold2 protein structures [60]. When applied to Arabidopsis seedlings exposed to excessive light, CysQuant successfully quantified the well-established increased reduction of Calvin-Benson cycle enzymes while also discovering previously uncharacterized redox-sensitive disulfides in chloroplastic enzymes [60]. These findings highlight how CysQuant enables comprehensive mapping of functionally relevant redox switches.
For drug development professionals, CysQuant offers a powerful approach for identifying and validating redox-dependent mechanisms in disease models. The methodology can be adapted to various mass spectrometry platforms, making it accessible to most proteomics facilities [60]. The versatility and precision of CysQuant make it particularly valuable for studying pathological conditions characterized by oxidative stress, where traditional proteomic approaches might miss crucial redox-dependent regulatory events.
DLF-Sul and the AI-Driven Prediction Landscape
Algorithmic Foundations and Predictive Features
DLF-Sul represents the cutting edge in computational prediction of cysteine sulfenylation, one of the fundamental oxidative modifications in redox signaling. As a machine learning or deep learning-based tool (specific architectural details from search results are limited), DLF-Sul builds on the established principles of successful prediction tools like pCysMod, which utilizes deep learning frameworks with particle swarm optimization for hyperparameter tuning [62]. These models typically extract multiple sequence-based features including binary encoding profiles, amino acid composition, position-specific scoring matrices, and composition of k-spaced amino acid pairs to represent the contextual environment around candidate cysteine residues [62].
The predictive power of these models stems from their ability to recognize patterns in protein sequences that correlate with redox susceptibility. Features such as local amino acid composition, solvent accessibility, evolutionary conservation, and charge distribution around cysteine residues contribute to the classification algorithms. For DLF-Sul specifically, the model is trained on experimentally verified sulfenylation sites, learning the complex sequence determinants that distinguish redox-sensitive cysteines from their non-reactive counterparts [53]. This training enables the tool to scan entire proteomes and prioritize candidate residues for experimental validation, dramatically accelerating the discovery process.
Research Applications and Integration with Experimental Workflows
The primary application of DLF-Sul lies in its ability to guide targeted redox proteomics studies. By predicting which cysteine residues are most likely to undergo sulfenylation, researchers can design focused experiments to validate these predictions using CysQuant or similar quantification methods. This integration of computational prediction with experimental validation creates a powerful feedback loop that continuously improves both the predictive models and our understanding of redox signaling networks.
In drug development contexts, DLF-Sul can identify novel redox-sensitive targets in disease-relevant pathways. For example, the tool could predict sulfenylation events in kinase signaling pathways that are known to be dysregulated in cancer or inflammatory diseases. These predictions can then inform the development of targeted covalent inhibitors that selectively interact with redox-sensitive cysteines, a approach exemplified by emerging redox-targeted covalent inhibitors (TCIs) [6]. The predictive guidance offered by DLF-Sul and similar tools thus accelerates both target discovery and therapeutic intervention strategies in redox-related pathologies.
Integrated Workflows: Bridging Prediction and Experimental Validation
The true power of computational tools like DLF-Sul emerges when they are integrated with experimental methodologies like CysQuant in unified workflows. This integration creates a synergistic cycle of discovery and validation that accelerates redox biology research.
Diagram 1: Integrated workflow bridging prediction and validation.
This integrated approach begins with computational prediction using DLF-Sul to scan proteomes of interest and generate prioritized lists of candidate redox-sensitive cysteine residues. These predictions then inform targeted experimental designs using CysQuant for validation. The resulting experimental data serves dual purposes: confirming biological findings and refining the predictive algorithms through iterative model improvement. This cyclic process continuously enhances both predictive accuracy and biological understanding.
For research professionals, implementing this integrated workflow requires careful experimental design. Key considerations include selecting appropriate biological models, establishing proper positive and negative controls, and determining optimal stimulation conditions to observe redox dynamics. The experimental protocol for such integrated studies typically involves:
- Sample Preparation: Cells or tissues are treated under relevant conditions (e.g., oxidative stress, receptor activation) and harvested using redox-preserving methods.
- Computational Prediction: DLF-Sul analysis of the proteome of interest generates candidate sulfenylation sites.
- CysQuant Processing: Samples undergo the CysQuant workflow including differential labeling, protein digestion, and mass spectrometry analysis.
- Data Integration: Experimental results from CysQuant are compared with computational predictions to validate targets and refine models.
- Functional Validation: Confirmed redox-sensitive proteins undergo further functional studies to determine biological significance.
This integrated approach maximizes resource efficiency by focusing experimental efforts on high-probability targets while simultaneously expanding the training data available for computational tool refinement.
Future Directions and Research Applications
The integration of AI-driven prediction tools like DLF-Sul with quantitative experimental methods like CysQuant is paving the way for transformative advances in redox biology and drug development. Several promising directions are emerging that will shape future research in this field.
First, the development of site-specific manipulation technologies represents a frontier in causal validation of redox signaling events. Emerging chemical biology strategies, such as integrating bioorthogonal cleavage chemistry with genetic code expansion, enable precise incorporation of sulfenic acid modifications at specific sites in proteins of interest [6]. Similarly, redox-targeted covalent inhibitors (TCIs) offer potential for therapeutic intervention by selectively blocking specific sulfenylation events in pathological contexts [6]. These approaches move beyond correlation to establish causation in redox signaling pathways.
Second, the application of these integrated approaches to disease-specific redoxome mapping holds tremendous promise for identifying novel therapeutic targets. For instance, comprehensive cysteine redoxome mapping in mouse liver models has already identified 169 cysteine residues with dynamic redox changes in response to high-fat/high-sucrose diet, mapping to 35 KEGG pathways central to redox balance and energy homeostasis [42]. Similar approaches in human disease models could reveal previously unappreciated redox-dependent mechanisms in cancer, metabolic disorders, and neurodegenerative diseases.
Finally, the continuing evolution of computational prediction algorithms will likely incorporate protein structural information from tools like AlphaFold2 to enhance prediction accuracy [60]. As structural data becomes more integrated with sequence-based features, predictive models will better account for the local electrostatic microenvironments and three-dimensional contexts that determine cysteine reactivity [42]. This structural awareness, combined with expanding experimental training data, will produce increasingly sophisticated tools for redox site prediction across diverse biological contexts.
For researchers and drug development professionals, these advances translate into more efficient target discovery, improved validation workflows, and novel therapeutic strategies targeting redox-sensitive pathways in human disease. The integration of computational prediction with experimental quantification represents nothing less than a paradigm shift in how we study and therapeutic target redox signaling networks.
Redox signaling, the biological process by which cells sense and respond to changes in redox state, operates largely through the reversible oxidation of reactive cysteine residues in proteins. These oxidative post-translational modifications (oxiPTMs) act as molecular switches that dynamically regulate protein function, localization, and interactions in response to metabolic and environmental cues [46] [40]. The integration of redox proteomics with transcriptomics and metabolomics represents a transformative approach in systems biology, enabling researchers to capture complex, multi-layer regulatory networks that control cellular responses in health and disease.
Cysteine residues undergo a variety of reversible oxidative modifications including S-sulfenylation (-SOH), S-glutathionylation (-SSG), S-nitrosylation (-SNO), and persulfidation (-SSH), each capable of altering protein structure and function [46] [6]. These modifications serve as critical sensors that link cellular redox status to broader physiological outputs. The recent advancement of mass spectrometry-based redox proteomics has enabled high-resolution mapping of these dynamic modifications, providing unprecedented insights into redox-regulated cellular processes [46] [63]. When combined with transcriptomic and metabolomic profiling, this integrated approach reveals how redox signals originating at the protein level propagate to influence gene expression and metabolic flux, offering a comprehensive view of cellular regulation.
Methodological Foundations: Experimental Workflows and Enrichment Strategies
Redox Proteomics Workflows
Comprehensive redox proteomics requires specialized workflows to capture and quantify the labile nature of cysteine oxiPTMs. The general workflow begins with careful sample preparation under controlled conditions to preserve the native redox state, followed by specific enrichment of redox-modified peptides, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, and computational processing for identification and quantification [46].
Table 1: Key Methodologies in Redox Proteomics
| Method | Principle | Applications | Considerations |
|---|---|---|---|
| Biotin-Switch Technique | Converts S-nitrosylated cysteines to biotin-tagged forms for enrichment | Selective detection of S-nitrosylation sites [46] | Requires strict control of lighting conditions; may have specificity limitations |
| Resin-Assisted Capture (RAC) | Thiol-affinity resin captures cysteine-containing peptides | Enrichment of various redox-modified cysteine peptides [46] | Compatible with multiple oxiPTM types; enables quantitative comparisons |
| Isotope-Coded Affinity Tags (ICAT) | Uses isotopically labeled tags to quantify oxidized vs. reduced cysteines | Quantitative redox proteomics; ratio-based quantification [46] | Provides site-specific quantification; can differentiate regulatory from stress-induced modifications |
| OxICAT | Isotope-coded affinity tag method specifically optimized for oxidation | Quantitative assessment of cysteine oxidation states [63] | Enables monitoring of localized cysteine oxidation in cells |
| IodoTMT | Uses isobaric tandem mass tags for multiplexed quantification | Multiplexed redox proteomics (up to 6-11 samples simultaneously) [46] | Enables high-throughput screening of redox dynamics across multiple conditions |
Transcriptomics and Metabolomics Integration
Transcriptomic profiling via RNA sequencing provides comprehensive data on gene expression patterns, while metabolomics and lipidomics characterize small molecules and lipid mediators that reflect cellular metabolic status. The power of multi-omics integration emerges from the simultaneous analysis of these complementary data layers, enabling the identification of regulatory relationships that would remain hidden in single-omics approaches [64] [65].
In a pioneering study investigating total-body irradiation in mice, researchers demonstrated this approach by combining RNA sequencing with mass spectrometry-based metabolomics and lipidomics of blood plasma. Their analysis revealed extensive radiation-induced alterations in both gene expression and metabolic pathways, with joint pathway analysis showing significant changes in amino acid, carbohydrate, lipid, nucleotide, and fatty acid metabolism [64]. This integrated methodology enabled the identification of key regulatory nodes connecting transcriptional changes to metabolic outcomes.
Computational Integration Strategies: Bridging Data Layers
The integration of multi-omics data requires sophisticated computational approaches that can identify significant correlations and interactions across different molecular layers. These methods range from correlation-based network analyses to advanced machine learning algorithms capable of identifying complex, non-linear relationships.
Correlation-Based Integration Methods
Correlation-based strategies identify statistical relationships between different types of omics data, revealing coordinated changes across molecular layers. These approaches include:
- Gene–Metabolite Network Analysis: Constructs correlation networks between gene expression and metabolite abundance patterns using Pearson correlation coefficients or similar measures. These networks visualize interactions between transcripts and metabolites, highlighting potential regulatory relationships [65].
- Weighted Correlation Network Analysis (WGCNA): Identifies modules of highly correlated genes and metabolites across samples, with eigengenes representing overall expression patterns for each module. Correlation between module eigengenes and metabolite patterns reveals metabolic pathways co-regulated with specific gene sets [65].
- Similarity Network Fusion: Constructs similarity networks for each omics data type separately, then merges them to identify edges with high associations across different omics layers, preserving strong multi-omics relationships [65].
Machine Learning and Advanced Computational Tools
Recent advances in artificial intelligence and machine learning have dramatically expanded capabilities for multi-omics integration. These approaches include:
- Predictive Modeling for Redox Modifications: Tools like CysQuant, BiGRUD-SA, DLF-Sul, and iCarPS utilize machine learning frameworks to predict redox-sensitive cysteine residues and their modifications based on protein sequence and structural features [46].
- Network-Based Integration: Platforms such as STITCH enable the construction of integrated networks showing interactions between proteins, metabolites, and other molecular species, revealing how redox modifications influence broader cellular networks [64].
- Joint-Pathway Analysis: Combined analysis of transcriptomic and metabolomic data within curated pathway databases identifies pathways showing significant alterations across multiple omics layers, highlighting coordinated biological responses [64].
Table 2: Computational Tools for Multi-Omics Integration
| Tool/Approach | Application | Key Features | Reference |
|---|---|---|---|
| WGCNA | Correlation network analysis | Identifies co-expressed gene modules correlated with metabolite patterns | [65] |
| CysQuant | Redox modification prediction | Machine learning-based prediction of redox-sensitive cysteine residues | [46] |
| STITCH | Network integration | Constructs protein-metabolite interaction networks | [64] |
| Similarity Network Fusion | Multi-omics data integration | Merges similarity networks from different omics data types | [65] |
| Joint-Pathway Analysis | Pathway enrichment | Identifies pathways altered across multiple omics layers | [64] |
Biological Applications: From Mechanisms to Medicine
Case Study: Radiation Response Mechanisms
The integrated multi-omics approach has been successfully applied to elucidate the complex biological responses to total-body irradiation. A landmark study combining transcriptomics with metabolomics and lipidomics in mice exposed to 1 Gy and 7.5 Gy radiation revealed:
- Dose-Dependent Molecular Responses: Clear separation of high-dose (7.5 Gy) groups from controls in multivariate analysis, with overlapping of low-dose (1 Gy) and control groups, demonstrating distinct molecular signatures at different exposure levels [64].
- Specific Molecular Alterations: Identification of dysregulated amino acids, various phosphatidylcholines (PC), phosphatidylethanolamines (PE), and carnitine derivatives, along with significantly dysregulated genes including Nos2, Hmgcs2, and Oxct2a [64].
- Integrated Pathway Analysis: Joint-Pathway Analysis revealed that radiation exposure resulted in coordinated changes in amino acid, carbohydrate, lipid, nucleotide, and fatty acid metabolism, with BioPAN analysis predicting key regulators (Elovl5, Elovl6, and Fads2) specifically in the high-dose group [64].
This integrated approach provided a systems-level understanding of radiation response mechanisms, demonstrating how redox proteomics combined with other omics layers can uncover complex physiological adaptations to environmental stressors.
Redox Signaling in Aging and Disease
Cysteine-mediated redox signaling plays a crucial role in the aging process and various age-related diseases. Quantitative characterization of reversible cysteine oxidation across ten mouse tissues revealed numerous tissue- and age-dependent changes in the cysteine redox proteome [40]. Multi-omics studies have shown that:
- Anti-aging Interventions: Dietary restriction, reduced insulin/IGF-1 signaling, and mTOR suppression all alter the cysteine redoxome in worms, flies, and rodents, suggesting a conserved mechanism for longevity assurance [40].
- Specific Redox-Regulated Proteins: An increasing number of aging-related proteins are modulated by redox reactions of cysteines, with specific oxidation events affecting organismal lifespan. For example, oxidation of a redox-sensitive cysteine in LET-60, a small GTPase regulating lifespan in nematodes, is required for longevity arising from increased ROS production [40].
- Metabolic Reprogramming: Integrated analyses reveal how redox modifications of metabolic enzymes create feedback loops that influence metabolic flux, gene expression, and ultimately cellular phenotypes in aging and disease [64] [40].
Successful integration of redox proteomics with other omics technologies requires specialized reagents and tools designed to capture, analyze, and interpret multi-omics data.
Table 3: Essential Research Reagents for Multi-Omics Redox Studies
| Reagent/Tool | Category | Function | Application Example |
|---|---|---|---|
| Iodoacetyl Tandem Mass Tags (iodoTMT) | Chemical Labeling | Multiplexed quantification of redox changes | Enabled identification of 70 redox-sensitive peptides during tomato fruit ripening [46] |
| Biotin-HPDP | Affinity Capture | Thiol-reactive biotin for pull-down assays | Forms disulfide bonds with reduced thiols in biotin-switch techniques |
| Dimedone-based Probes | Chemical Probes | Selective detection of sulfenic acid modifications | Early tools for SOH detection; foundation for targeted covalent inhibitors [6] |
| TurboID/APEX | Proximity Labeling | Enable monitoring of localized cysteine oxidation | Combined with OxICAT for spatial redox proteomics [63] |
| N-ethylmaleimide (NEM) | Alkylating Agent | Blocks free thiols to prevent post-collection oxidation | Standard in redox proteomics workflows to preserve native oxidation states |
| CysQuant | Computational Tool | Predicts redox-sensitive cysteine residues | Machine learning-based prediction of oxidative modifications [46] |
Future Perspectives and Concluding Remarks
The integration of redox proteomics with transcriptomics and metabolomics represents a powerful paradigm for understanding complex biological systems. As these technologies continue to evolve, several emerging trends are poised to further transform the field:
- Precision Manipulation of Redox Signaling: Emerging chemical biology strategies enable site-specific manipulation of cysteine oxidation, including bioorthogonal cleavage chemistry combined with genetic code expansion for controlled activation of redox events, and targeted covalent inhibitors to selectively block specific SOH modifications [6].
- Single-Cell Multi-Omics: Advances in single-cell technologies will enable the investigation of redox regulation with unprecedented resolution, revealing cell-type-specific redox signaling networks and their functional consequences in complex tissues [65].
- AI-Driven Integration: Artificial intelligence and machine learning algorithms are increasingly capable of identifying complex, non-linear relationships across omics layers, predicting functional outcomes from integrated redox proteomic, transcriptomic, and metabolomic data [46] [66].
- Clinical Translation: Integration of multi-omics data holds significant promise for identifying novel therapeutic targets and biomarkers for diseases characterized by redox dysregulation, including cancer, neurodegenerative disorders, and metabolic diseases [64] [66].
The continued refinement and integration of these approaches will undoubtedly yield new insights into the fundamental principles of redox biology and provide novel therapeutic avenues for manipulating redox signaling in disease contexts. As these technologies become more accessible and computational methods more sophisticated, the comprehensive mapping of redox-regulated networks across multiple biological layers will become increasingly routine, driving innovation in both basic research and clinical applications.
The cysteine redoxome represents the comprehensive landscape of reversible oxidative post-translational modifications (PTMs) on protein cysteine residues, serving as molecular "switches" that regulate protein function, signaling pathways, and metabolic homeostasis [67] [68]. In metabolic diseases such as type 2 diabetes and metabolic dysfunction-associated steatotic liver disease (MASLD), dysregulation of redox balance disrupts normal cysteine switching behavior, contributing to pathological states [67] [68]. This case study examines the application of cysteine redoxome mapping to elucidate the molecular mechanisms underlying hepatic metabolic dysfunction, providing a technical framework for investigating redox regulation in disease contexts.
The functional versatility of cysteine stems from the sulfur atom in its thiol group, which can adopt multiple oxidation states in response to redox fluctuations [69] [40]. Under physiological conditions, reactive oxygen species (ROS) and hydrogen sulfide (Hâ‚‚S) mediate reversible cysteine modifications including S-sulfenylation (-SOH), S-nitrosylation (-SNO), S-glutathionylation (-SSG), and disulfide bond formation (-S-S-) [21] [40]. These modifications dynamically regulate protein activity, subcellular localization, and interaction networks, enabling cells to adapt to metabolic and oxidative challenges [40].
Within the context of a broader thesis on redox regulation, this case study demonstrates how comprehensive cysteine redoxome profiling provides unprecedented insights into the molecular pathology of metabolic liver disease, revealing novel therapeutic targets and biomarker candidates for drug development.
Technical Foundations of Cysteine Redoxome Mapping
Fundamental Principles of Cysteine Redox Chemistry
Cysteine residues function as sentinels of cellular redox status due to the unique reactivity of their thiol groups. The sulfur atom's d-orbital electrons enable transitions between reduced (thiol, -SH) and multiple oxidized states, forming a redox code that integrates information from various reactive species including H₂O₂, nitric oxide (•NO), and H₂S [40] [70]. This chemical versatility allows cysteine residues to act as binary switches that regulate protein structure and function in response to redox challenges [68].
The reactivity of specific cysteine residues is governed by their electrostatic microenvironments. Positively charged amino acids (e.g., lysine, arginine) near cysteine residues lower the pKa of the thiol group, stabilizing the thiolate anion (-Sâ») form that is highly susceptible to oxidation [69] [68]. Conversely, negatively charged residues (e.g., aspartate, glutamate) increase thiol pKa, reducing redox sensitivity [69]. This principle enables prediction of redox-sensitive cysteines based on primary protein sequences and three-dimensional structures.
Experimental Workflow for Comprehensive Redoxome Assessment
The following diagram illustrates the core experimental workflow for cysteine redoxome mapping using differential alkylation-based proteomics:
Figure 1: Experimental workflow for cysteine redoxome mapping using differential alkylation and quantitative proteomics.
Key Methodological Considerations
Sample Preparation: Rapid processing under controlled conditions is essential to preserve native redox states. Lysis buffers typically include alkylating agents (e.g., N-ethylmaleimide [NEM]) to block free thiols and prevent artificial oxidation during extraction [68]. Protease and phosphatase inhibitors maintain protein integrity, while mechanical disruption ensures complete lysis.
Differential Alkylation: This cornerstone technique involves sequential labeling of reduced and oxidized cysteine pools [68]. Initially, reduced cysteines are blocked with NEM. Subsequently, reversibly oxidized cysteines are reduced with dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP) and labeled with a distinct alkylating agent such as biotin-PEAC5-maleimide (BPM) or isotope-coded iodoacetamide (IAM) [68] [50]. This approach enables simultaneous quantification of reduced and oxidized cysteine residues.
Mass Spectrometry Analysis: High-resolution LC-MS/MS systems (e.g., Orbitrap platforms) provide the sensitivity and dynamic range needed for proteome-wide cysteine quantification [50]. Advanced fragmentation techniques (MS3) improve peptide identification, while isobaric labeling (e.g., TMT, iodoTMT) enables multiplexed analysis of multiple conditions [71].
Hepatic Cysteine Redoxome in Metabolic Disease: A Representative Study
Experimental Model and Design
A recent investigation applied cysteine redoxome mapping to examine the effects of a high-fat/high-sucrose diet (HFHSD) on hepatic redox regulation in male mice [67] [68]. The study employed C57BL/6 mice fed either a normal chow diet (NCD) or HFHSD for 16 weeks (n=4 per group), inducing obesity, insulin resistance, and hepatic steatosis reminiscent of human metabolic disease [68].
Liver tissues were processed using a differential alkylation protocol wherein reduced cysteines were labeled with NEM, reversibly oxidized cysteines were reduced with DTT and labeled with BPM, and samples were subjected to tryptic digestion [68]. Peptides were analyzed by LC-MS/MS, with data processing using MaxQuant and statistical analysis in Perseus [50].
Global Redoxome and Proteome Alterations
The comprehensive analysis identified and quantified over 5,000 cysteine residues in both reduced and oxidized states [68]. Surprisingly, HFHSD did not induce widespread shifts in the global cysteine redox equilibrium, with similar distributions of oxidized and reduced cysteine residues across subcellular compartments in both diet groups [67] [68].
Table 1: Proteomic alterations in HFHSD mouse livers
| Category | Upregulated Processes/Pathways | Downregulated Processes/Pathways |
|---|---|---|
| Biological Processes | Nucleosome assembly, nucleosome organization, protein localization to chromatin | Unsaturated fatty acid metabolic process, carboxylic acid metabolic process, organic acid metabolic process |
| KEGG Pathways | Oxidative phosphorylation, thermogenesis, nonalcoholic fatty liver disease, PPAR signaling pathway | Steroid hormone biosynthesis, retinol metabolism, linoleic acid metabolism, arachidonic acid metabolism |
| Functional Implications | Genomic stability, lipid detoxification, energy regulation | Detoxification capacity, metabolic flexibility |
Parallel proteomic analysis revealed that HFHSD upregulated 475 proteins involved in genomic stability, lipid detoxification, and energy regulation, while downregulating 208 proteins linked to detoxification and metabolic flexibility [68]. This proteomic remodeling indicates hepatic adaptation to metabolic stress, with preserved cysteine redox homeostasis despite significant protein expression changes.
Dynamic Cysteine Residues and Pathway Analysis
Despite global redox stability, the study identified 169 cysteine residues exhibiting significant redox changes in response to HFHSD, mapping to 35 KEGG pathways central to redox balance and energy homeostasis [67] [68]. These dynamic cysteines represent sensitive nodes within the hepatic redox network that potentially drive metabolic dysfunction.
Table 2: Characteristics of HFHSD-sensitive cysteine residues
| Feature | Oxidation-Sensitive Cysteines | Reduction-Sensitive Cysteines |
|---|---|---|
| Subcellular Enrichment | Mitochondria and cytosol | Extracellular regions |
| Structural Preference | Buried within protein structures | Exposed on protein surfaces |
| Functional Roles | Catalytic activity, regulatory switches | Disulfide bond formation, molecular switches |
| Electrostatic Microenvironment | Positively charged amino acids | Varied, context-dependent |
| Representative Proteins | Metabolic enzymes, redox sensors | Secreted proteins, membrane receptors |
Structural and motif analyses revealed that HFHSD-induced oxidation-sensitive cysteines were enriched in mitochondria and cytosol, while reduction-sensitive cysteines localized predominantly to extracellular regions [68]. This compartmentalization suggests distinct regulatory mechanisms operating in different subcellular environments.
Analytical Approaches and Bioinformatics Strategies
Cysteine Redoxome Data Analysis Pipeline
The complexity of cysteine redoxome datasets requires specialized bioinformatic approaches. The following diagram outlines a comprehensive data analysis pipeline:
Figure 2: Bioinformatics pipeline for cysteine redoxome data analysis.
Key Bioinformatics Tools and Techniques
Quality Control and Filtering: Stringent criteria exclude low-confidence identifications, typically requiring cysteine quantification in at least 2-3 replicates with coefficient of variation (CV) <20% between replicates [68] [50]. These measures ensure data reliability despite the technical challenges of redox proteomics.
Statistical Analysis: The Significance B algorithm in Perseus identifies significantly altered cysteine residues, accounting for intensity-dependent variance in quantification accuracy [50]. Multiple testing correction (e.g., Benjamini-Hochberg) controls false discovery rates.
Pathway and Network Analysis: Enrichment analysis using Gene Ontology (GO) and KEGG databases identifies biological processes and pathways enriched with dynamic cysteines [67] [68]. Integration with protein-protein interaction networks (e.g., STRING) reveals redox-sensitive modules within cellular signaling networks.
Motif and Structural Analysis: Sequence alignment tools (e.g., MUSCLE) assess cysteine conservation across species [69]. Motif analysis (pLogo) identifies amino acid preferences around redox-sensitive cysteines, while structural modeling predicts solvent accessibility and electrostatic environments [68].
Research Reagent Solutions for Cysteine Redoxome Studies
Table 3: Essential research reagents for cysteine redoxome mapping
| Reagent Category | Specific Examples | Function & Application |
|---|---|---|
| Thiol-Alkylating Agents | N-ethylmaleimide (NEM), iodoacetamide (IAM), biotin-PEAC5-maleimide (BPM) | Block free thiols, label reduced/oxidized cysteine pools |
| Reducing Agents | Dithiothreitol (DTT), tris(2-carboxyethyl)phosphine (TCEP) | Reduce reversible oxidative modifications (disulfides, sulfenylation) |
| Isobaric Tags | iodoTMT, TMT | Multiplexed quantification of cysteine oxidation across samples |
| Affinity Tags | Biotin-maleimide, Azide-alkyne handles | Enrich cysteine-containing peptides for comprehensive coverage |
| Protease Inhibitors | PMSF, protease inhibitor cocktails | Prevent protein degradation during sample processing |
| Antibodies for Validation | Anti-biotin, anti-sulfenylation (e.g., dimedone-based) | Confirm specific cysteine modifications via immunoblotting |
Integration with Redox Signaling Pathways in Metabolic Disease
The hepatic cysteine redoxome functions as an integrative interface between metabolic pathways and redox signaling networks. The following diagram illustrates key redox-sensitive pathways in metabolic liver disease:
Figure 3: Redox signaling pathways in HFHSD-induced metabolic liver disease.
Regulatory Mechanisms and Adaptive Responses
Sulfiredoxin-1 (SRXN1) in Redox Homeostasis: SRXN1 has emerged as a key regulator of protein sulfinylation, particularly in reducing cysteine-sulfinic acid (Cys-SOâ‚‚H) modifications on peroxiredoxins and other target proteins [21]. SRXN1 expression is transcriptionally regulated by Nrf2 and AP-1, creating a feedback loop that fine-tunes cellular responses to oxidative stress [21]. In metabolic liver disease, SRXN1-mediated repair of sulfinylated proteins represents a crucial adaptive mechanism against chronic oxidative challenge.
Cross-talk Between ROS and Hâ‚‚S Signaling: Physiological ROS and Hâ‚‚S signaling are intrinsically connected through cysteine modification networks [40]. Hydrogen peroxide-mediated sulfenylation of specific cysteines (e.g., Cys797 in EGFR) enhances kinase activity, while Hâ‚‚S promotes persulfidation that can antagonize ROS signaling [40]. This yin-yang relationship between oxidizing and reducing species creates a sophisticated regulatory layer for metabolic control.
Redox Control of Metabolic Enzymes: HFHSD-sensitive cysteine residues identified in redoxome studies are enriched in metabolic pathways including fatty acid oxidation, tricarboxylic acid cycle, and oxidative phosphorylation [67] [68]. Reversible oxidation of catalytic or allosteric cysteines in metabolic enzymes enables rapid adjustment of flux through metabolic pathways in response to nutritional status and cellular energy demands.
This case study demonstrates how comprehensive cysteine redoxome mapping provides unprecedented insights into the molecular pathology of metabolic liver disease. The application of differential alkylation coupled with quantitative proteomics has revealed that HFHSD induces selective redox remodeling of specific cysteine residues despite global redox homeostasis, identifying vulnerable nodes within hepatic signaling and metabolic networks.
For drug development professionals, the cysteine redoxome represents a rich landscape of potential therapeutic targets. Small molecules that selectively modulate the redox state of key cysteine residues could offer novel approaches for treating metabolic diseases [21]. Additionally, circulating proteins with redox-sensitive cysteines may serve as biomarkers for monitoring disease progression and therapeutic responses [50].
Future directions in cysteine redoxome research will likely focus on single-cell redox profiling, spatial redoxomics within tissue contexts, and dynamic monitoring of cysteine oxidation in living systems [70]. Integration of redox proteomics with other omics technologies will provide systems-level understanding of redox regulation, potentially revealing new therapeutic avenues for metabolic and other redox-related diseases.
Overcoming Specificity and Translational Challenges in Redox Research
The redox regulation of protein cysteine residues represents a fundamental signaling mechanism in biology, governing processes from plant stress responses to mammalian aging and disease. Oxidative post-translational modifications (oxiPTMs) function as molecular switches that dynamically alter protein function, structure, and interactions in response to cellular redox changes [46] [47]. Unlike stable protein modifications, oxiPTMs such as S-sulfenylation (-SOH), S-nitrosation (-SNO), S-glutathionylation (-SSG), and persulfidation (-SSH) are notably transient and reversible, with lifetimes ranging from milliseconds to seconds under physiological conditions [48] [72]. This inherent lability presents substantial methodological challenges for their comprehensive detection, characterization, and functional analysis.
The reactivity of cysteine residues stems from the nucleophilic property of the thiolate anion (-S−), which is susceptible to oxidation by reactive oxygen, nitrogen, and sulfur species (ROS/RNS/RSS) [48]. The resulting modifications exist in a dynamic equilibrium, where sulfenic acid often serves as a pivotal intermediate that can rapidly evolve into more stable forms through reactions with other thiols or cellular metabolites [6] [72]. This technical whitepaper synthesizes contemporary strategies for capturing and stabilizing these elusive oxiPTMs, providing researchers with experimentally validated methodologies to advance the field of cysteine-mediated redox signaling.
Fundamental Chemistry of Cysteine OxiPTMs and Their Interconversions
Understanding the chemical landscape of cysteine modifications is prerequisite to developing effective capture strategies. The oxidation state of sulfur in oxiPTMs spans a wide range from −2 to +6, with distinct biological implications for each form [48]. The initial reaction of a protein thiol with hydrogen peroxide forms sulfenic acid, a crucial signaling intermediate that can subsequently follow multiple chemical trajectories:
- Reaction with glutathione to form S-glutathionylation
- Formation of intra- or inter-molecular disulfide bonds
- Further oxidation to sulfinic and irreversible sulfonic acids
- Reaction with hydrogen sulfide to form persulfidation [40] [48] [72]
This complex network of potential interconversions necessitates highly specific stabilization approaches tailored to each oxiPTM type. The thermodynamics and kinetics of these reactions are profoundly influenced by the cysteine's microenvironment, particularly its pKa and the electrostatic properties of the surrounding residues [72]. Cysteines with lowered pKa values exist as thiolate anions at physiological pH, significantly enhancing their nucleophilicity and consequent susceptibility to oxidation [48]. This chemical understanding directly informs the design of targeted capture reagents that exploit the unique reactivity profiles of each oxiPTM.
OxiPTM Interconversion Pathways
Figure 1: Cysteine OxiPTM Interconversion Pathways. The diagram illustrates the dynamic equilibrium between different oxidative post-translational modifications of cysteine residues, highlighting sulfenic acid as a central intermediate. Solid arrows represent chemical conversions, while the dashed arrow indicates enzymatic reduction. Coloring distinguishes reduction states: green (reduced), yellow (key intermediate), red (reversible oxiPTMs), and blue (partially or fully irreversible oxidations).
Chemoproteomic Strategies for Specific OxiPTM Capture
Modern chemoproteomics employs targeted chemical probes that react specifically with different oxiPTM forms, enabling their selective enrichment and subsequent mass spectrometry analysis. These approaches have revolutionized our ability to profile the "redoxome" – the comprehensive repertoire of oxidized cysteine residues within a biological system [48].
Sulfenic Acid (-SOH) Capture Using Nucleophilic Probes
Sulfenic acids are highly reactive intermediates that can be selectively trapped using carbon nucleophiles. The cyclic 1,3-diketone dimedone and its derivatives form stable thioether adducts with -SOH, enabling specific labeling without reacting with other oxiPTMs [48] [6]. Contemporary probes build upon this chemistry with enhanced functionality:
- DYn-2 (4-(pent-4-yn-1-yl)cyclohexane-1,3-dione): An alkyne-functionalized dimedone derivative that enables click chemistry-based biotin conjugation for streptavidin enrichment [48]
- BTD-based probes: Benzothiazine derivatives offering improved cell permeability and reaction kinetics [72]
- Photoaffinity dimedone analogs: Incorporating photochemically reactive groups for covalent crosslinking
The application of DYn-2 in Arabidopsis thaliana cell cultures identified 1,394 S-sulfenylated proteins, demonstrating the remarkable scope of this modification in plant redox signaling [72]. Similar approaches in human cells have revealed sulfenylation networks in growth factor signaling pathways [73].
S-Nitrosation (-SNO) Detection via the Biotin-Switch Technique
The Biotin-Switch Technique (BST) remains the cornerstone methodology for S-nitrosation detection, employing a three-step process:
- Block free thiols with methyl methanethiosulfonate (MMTS) or N-ethylmaleimide (NEM)
- Selectively reduce S-NO bonds with ascorbate or copper(II)/cysteine
- Label newly revealed thiols with biotin-HPDP for affinity purification [48] [74]
Modified BST protocols using iodoTMT tags enable multiplexed quantification of S-nitrosated peptides across different experimental conditions [46] [72]. In Arabidopsis guard cells, this approach identified 35 S-nitrosated proteins responsive to flg22 treatment, illustrating the technique's applicability to specific cell types and signaling contexts [72].
Persulfidation (-SSH) and S-Glutathionylation (-SSG) Profiling
Persulfidation detection employs a modified BST wherein -SSH residues are selectively reduced by dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP) after blocking free thiols and S-nitrosated sites [48] [72]. The resulting thiols are then captured with biotin-based reagents. This approach identified 106 persulfidated proteins in Arabidopsis leaves, expanding the recognized scope of hydrogen sulfide signaling [72].
S-glutathionylation profiling leverages biotinylated glutathione ethyl ester (BioGEE), which cells incorporate into their glutathione pool. Under oxidative stress, BioGEE forms adducts with target proteins that can be affinity-purified and identified by MS. Alternative approaches use anti-glutathione antibodies for immunocapture, though with potentially lower specificity [72].
Table 1: Quantitative Profiling of OxiPTMs in Plant Systems
| OxiPTM | Number of Identified Proteins | Biological System | Primary Method | Reference |
|---|---|---|---|---|
| S-sulfenylation | 1,394 | Arabidopsis cell culture + Hâ‚‚Oâ‚‚ | BTD-based probe | [72] |
| S-sulfenylation | 132 | Arabidopsis plastids | YAP1C probe | [72] |
| S-glutathionylation | 79 | Arabidopsis cell culture | GS-biotin labeling + 2D-PAGE/MALDI-MS | [72] |
| S-nitrosation | 926 | Arabidopsis | Site-specific nitrosoproteomics | [72] |
| S-nitrosation | 35 | Arabidopsis guard cells + flg22 | IodoTMT labeling | [72] |
| Persulfidation | 106 | Arabidopsis leaves | Modified Biotin-Switch Technique | [72] |
| S-cyanylation | 163 | Arabidopsis leaves | Chemoproteomic profiling | [72] |
Experimental Workflows for Redoxome Profiling
Comprehensive redoxome analysis requires integrated workflows that maintain the native oxidation state of cysteines throughout sample processing while enabling specific enrichment of modified residues.
Quantitative Thiol Reactivity Profiling (QTRP)
QTRP assesses the intrinsic reactivity of cysteine residues by treating native proteomes with iodoacetamide-alkyne (IA) or 2-iodo-N-(prop-2-yn-1-yl)acetamide (IPM) probes at varying concentrations [73] [48]. The fundamental principle states that hyperreactive cysteines saturate labeling at low probe concentrations, while less reactive thiols exhibit concentration-dependent labeling. This approach was applied to C. elegans, quantifying reactivity for 5,258 cysteine sites and identifying 1,292 hyperreactive residues likely to have functional significance [73].
Protocol: QTRP for Reactivity Assessment
- Prepare fresh lysates under non-denaturing conditions with minimal oxygen exposure
- Treat parallel samples with 10µM and 100µM IPM probe for 1 hour at room temperature
- Perform click chemistry with light or heavy isotope-coded Az-UV-biotin tags
- Enrich biotinylated peptides using streptavidin chromatography
- Elute via UV cleavage and analyze by LC-MS/MS
- Calculate R100:10 ratio (labeling at 100µM vs 10µM) to determine reactivity [73]
Resin-Assisted Capture (RAC) of Reversible OxiPTMs
RAC provides a versatile platform for enriching various reversible oxiPTMs through thiol-disulfide exchange chemistry. The general workflow involves:
- Blocking free thiols with N-ethylmaleimide (NEM) or iodoacetamide (IAM)
- Selective reduction of target oxiPTMs:
- Ascorbate for S-nitrosation
- DTT/TCEP for disulfides and persulfidation
- Glutaredoxin/thioredoxin systems for specific disulfide bonds
- Capture of newly reduced thiols on thiol-affinity resins like Thiopropyl Sepharose
- Elution and identification of captured proteins [46] [48]
RAC has been successfully applied in plant systems, such as identifying redox-sensitive proteins in tomato fruit ripening where 70 redox-sensitive peptides from 51 proteins were quantified using iodoTMT-based RAC [46].
OxiPTM Workflow Integration
Figure 2: Integrated Workflows for OxiPTM Capture and Enrichment. The diagram outlines two complementary approaches for comprehensive redoxome profiling. Reductomic methods (upper path) rely on selective reduction and capture, while chemoproteomic approaches (lower path) utilize targeted chemical probes for specific oxiPTMs. Both pathways converge on LC-MS/MS for identification and quantification.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Essential Research Reagents for OxiPTM Capture and Analysis
| Reagent Category | Specific Examples | Function | Applications |
|---|---|---|---|
| Thiol Blocking Reagents | N-ethylmaleimide (NEM), Iodoacetamide (IAM), Methyl methanethiosulfonate (MMTS) | Irreversibly alkylate free thiols to prevent artifactual oxidation | Sample preparation prior to specific oxiPTM reduction |
| Sulfenic Acid Probes | Dimedone, DYn-2, BTD-based probes | Chemoselectively label -SOH forms via nucleophilic addition | Sulfenylation profiling in cells and tissues |
| Alkyne-Tagged Probes | IAM-alkyne (IA), IPM probe | Enable bioorthogonal conjugation via click chemistry | QTRP, enrichment for MS analysis |
| Biotin Switching Reagents | Biotin-HPDP, N-[6-(biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide | Thiol-reactive biotinylation after selective reduction | BST for S-nitrosation, persulfidation |
| Affinity Enrichment Matrices | Streptavidin beads, Thiopropyl Sepharose | Capture biotinylated or thiol-containing proteins/peptides | Enrichment of modified cysteines from complex mixtures |
| Isotope Tags | IodoTMT, ICAT (Isotope-Coded Affinity Tags) | Enable multiplexed quantification of oxiPTMs | Comparative redoxomics across conditions |
| Selective Reducing Agents | Ascorbate, DTT, TCEP, Arsenite | Specifically reduce target oxiPTMs while preserving others | Differential reduction in RAC workflows |
| 3-Methyl-6-nitro-1H-indazole | 3-Methyl-6-nitro-1H-indazole | Research Chemical | High-purity 3-Methyl-6-nitro-1H-indazole for research applications. For Research Use Only. Not for human or veterinary use. | Bench Chemicals |
| Avn-492 | Avn-492, MF:C17H21N5O2S, MW:359.4 g/mol | Chemical Reagent | Bench Chemicals |
Advanced Techniques for Site-Specific Manipulation
Genetic Code Expansion for Controlled SOH Formation
Traditional genetic approaches (cysteine-to-serine mutagenesis) disrupt all cysteine functions, including structural roles and metal binding. Emerging strategies now enable precise, site-specific incorporation of sulfenic acid modifications using genetic code expansion technology:
- Incorporation of photocaged cysteine sulfoxides as unnatural amino acids (UAAs)
- Controlled activation via UV irradiation to generate SOH on demand
- Functional assessment without global redox perturbation [6]
This approach utilizes DMNB-caged cysteine sulfoxide, which is incorporated into proteins of interest via an orthogonal synthetase/tRNA pair. Subsequent UV illumination cleaves the photolabile cage, generating authentic sulfenic acid at defined positions. Initial proof-of-concept studies demonstrated this technology with peroxidase Gpx3, enabling controlled SOH formation and facilitating functional studies of specific redox events [6].
Targeted Covalent Inhibitors for Redox Signaling
The success of covalent kinase inhibitors (e.g., afatinib, ibrutinib) inspires the development of targeted covalent inhibitors (TCIs) for redox signaling. These compounds consist of:
- A target-binding module for specificity
- A moderately reactive warhead (e.g., nitroacetamide) for covalent modification
- Optimization for selective engagement with redox-active cysteines [6]
Early examples include dimedone-conjugated compounds targeting protein tyrosine phosphatases, though these initial designs lack sufficient proteome-wide specificity. Next-generation redox TCIs aim to achieve precise inhibition of specific SOH-mediated signaling events without global antioxidant effects, potentially offering therapeutic applications in conditions with dysregulated redox signaling [6].
The field of cysteine redoxomics has progressed from merely detecting oxidative modifications to precisely manipulating specific redox events within complex biological systems. The strategies outlined—from chemoselective probes to genetic code expansion—provide researchers with an expanding toolkit to address the transient nature of oxiPTMs. As these methodologies continue to evolve, they will undoubtedly uncover deeper insights into redox signaling networks and their roles in health and disease.
Future directions will likely focus on increasing temporal resolution for capturing rapid redox dynamics, enhancing spatial mapping within subcellular compartments, and developing more sophisticated computational models to predict redox-sensitive cysteines and their functional outcomes [46]. The integration of these advanced capture and stabilization strategies with multi-omics approaches will further establish redox proteomics as a cornerstone of systems biology, enabling unprecedented understanding of cysteine-mediated redox regulation in physiological and pathological contexts.
Cysteine is one of the least abundant yet most highly conserved amino acids in proteins, performing diverse functional roles including catalytic activity, metal binding, and redox regulation [75]. Its remarkable chemical versatility stems from the sulfur atom in its thiol group, which is highly nucleophilic and sensitive to oxidative modification [76] [75]. Unlike other amino acids, functional cysteines do not conform to a canonical sequence motif, complicating their systematic identification and characterization [76]. Instead, cysteine reactivity is profoundly shaped by two key determinants: the immediate electrostatic microenvironment and the broader subcellular localization. This review examines how these factors collectively tune cysteine reactivity within the context of redox regulation, providing a framework for understanding cysteine-mediated signaling in health and disease.
The Electrostatic Microenvironment: Fine-Tuning Cysteine Reactivity
The intrinsic nucleophilicity of a cysteine residue is primarily governed by the ionization state of its thiol group, which is highly sensitive to the local electrostatic environment surrounding the residue [77] [76].
Positively Charged Residues Enhance Reactivity
The reactivity of cysteine thiols is profoundly influenced by neighboring amino acid residues. Research on rat brain tubulin revealed that cysteines exhibiting rapid reactivity with sulfhydryl reagents (Cys-305α, Cys-315α, Cys-316α, Cys-347α, Cys-376α, Cys-241β, and Cys-356β) were consistently located within 6.5 Å of positively charged residues such as arginine and lysine, or the positive edges of aromatic rings [77]. These positive charges presumably promote the dissociation of the thiol (-SH) to the more nucleophilic thiolate anion (-S-) at physiological pH, thereby enhancing reactivity [77]. Conversely, the inactivity of less reactive cysteines was ascribed to negatively charged local environments that suppress thiolate formation, even when these cysteines were surface-exposed [77].
Quantifying Hyperreactivity and Functional Significance
Hyperreactivity is a rare feature among cysteines that strongly predicts functional importance. Using the isoTOP-ABPP (isotopic Tandem Orthogonal Proteolysis – Activity-Based Protein Profiling) method, researchers can quantitatively profile cysteine reactivity proteome-wide [76]. This approach involves labeling cysteine residues with an iodoacetamide (IA) probe containing an alkyne handle, followed by click chemistry conjugation to azide-functionalized TEV-protease recognition peptides with isotopic labels for quantitative mass spectrometry analysis [76].
Table 1: Functional Annotation of Cysteines by Reactivity Profile
| Reactivity Category | isoTOP-ABPP Ratio (R[high]:[low]) | Prevalence | Enrichment in Functional Residues | Examples |
|---|---|---|---|---|
| Hyperreactive | < 2 | < 10% of cysteines | ~35% (active-site nucleophiles, redox-active disulfides) | C32 in GSTO1, C126 in ACAT1 |
| Less Reactive | >> 1 | ~90% of cysteines | ~0.2% (background level) | C90, C192, C237 in GSTO1 |
This quantitative profiling reveals that hyperreactive cysteines are remarkably enriched in functional residues, with 35% of cysteines with R10:1 < 2 being annotated as active-site nucleophiles or redox-active disulfides compared to just 0.2% for all cysteine residues in the UniProt database [76]. For example, in glutathione S-transferase GSTO1, among four cysteine residues labeled, only C32—the known active-site nucleophile—displayed hyperreactivity (R10:1 = 0.9), while the other three cysteines showed significantly higher ratios indicating lower reactivity [76].
Subcellular Localization: Compartmentalized Control of Cysteine Function
Beyond the immediate protein microenvironment, the physicochemical conditions of subcellular organelles create distinct landscapes that shape cysteine reactivity and function [75].
Organelle-Specific pH and Redox Potentials
Eukaryotic organelles maintain distinct pH and reduction potentials that significantly influence cysteine reactivity:
Table 2: Physicochemical Conditions of Mammalian Subcellular Organelles
| Organelle | pH | Redox Potential (GSH/GSSG) | Impact on Cysteine Reactivity |
|---|---|---|---|
| Cytosol | ~7.2 | -220 to -260 mV | Slightly favors thiolate formation |
| Mitochondrial Matrix | ~8.0 | -300 to -330 mV | Alkaline pH stabilizes thiolate; highly reducing environment primes cysteines for redox signaling |
| Endoplasmic Reticulum | ~7.2 | -150 mV | Oxidizing environment promotes structural disulfide bond formation |
| Lysosome | 4.5-5.0 | Not specified | Acidic pH favors protonated thiol; specialized enzymes (cathepsins) have depressed pKa values |
The mitochondrial matrix, with its alkaline pH (8.0) and highly reducing environment (-300 to -330 mV), theoretically stabilizes thiolates to produce more reactive cysteine residues [75]. This environment is essential for mitochondrial-specific cysteine functions, including iron-sulfur cluster biosynthesis and cysteine persulfide formation [75]. Conversely, the oxidizing environment of the endoplasmic reticulum (-150 mV) supports the formation and retention of structural disulfide bonds on secretory proteins [75].
Organelle-Specific Cysteine Functions
Mitochondria: Iron-Sulfur Clusters and Persulfide Signaling
Mitochondria harbor unique cysteine-dependent processes, most notably the biosynthesis of cellular iron-sulfur (Fe-S) clusters [75]. The assembly of [2Fe-2S] clusters on the scaffold protein ISU1 requires cysteine residues for cluster ligation and transfer [75]. These cysteine ligands must remain reduced before Fe-S cluster insertion, facilitated by the highly reducing conditions of the mitochondrial matrix [75].
Additionally, mitochondria are central to cysteine persulfide (-SSH) formation through enzymatic activities such as mercaptopyruvate sulfur transferase (MST) and sulfide:quinone oxidoreductase (SQR) in the hydrogen sulfide (Hâ‚‚S) oxidation pathway [75]. These persulfide modifications represent an emerging layer of redox regulation mediated by specialized cysteine residues within this compartment.
Endoplasmic Reticulum: Oxidative Protein Folding
The oxidizing environment of the endoplasmic reticulum (-150 mV) supports the formation of structural disulfide bonds, which are essential for proper protein folding and stability of secretory proteins [75]. This process involves protein disulfide isomerases (PDIs) and oxidoreductases such as ERO1, which generate Hâ‚‚Oâ‚‚ as a byproduct during disulfide bond formation [40].
Experimental Methods for Profiling Cysteine Reactivity
The isoTOP-ABPP Workflow
The isotopic Tandem Orthogonal Proteolysis – Activity-Based Protein Profiling (isoTOP-ABPP) method enables quantitative, proteome-wide analysis of cysteine reactivity in native biological systems [76]. The detailed methodology consists of several key stages:
- Probe Labeling: Native proteomes are treated with low concentrations (micromolar) of an iodoacetamide (IA) probe functionalized with an alkyne handle. This probe covalently modifies reactive cysteine residues.
- Click Chemistry: Probe-labeled proteins are conjugated via copper-catalyzed azide-alkyne cycloaddition ("click chemistry") to an azide-functionalized TEV protease recognition peptide containing a biotin affinity handle and an isotopic label (light or heavy valine).
- Affinity Enrichment and Proteolysis: Labeled proteins are captured on streptavidin beads and undergo tandem on-bead proteolytic digestions: first with trypsin, then with TEV protease, which specifically releases the probe-labeled peptides.
- LC-MS/MS Analysis: The isotopically labeled, probe-modified peptides are analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The MS2 spectra identify the specific cysteine-containing peptide and modification site, while the MS1 spectra enable quantification of the relative abundance (and thus reactivity) of each cysteine based on the light-to-heavy isotope ratio.
Subcellular Organelle Targeting Strategies
Recent advancements in chemoproteomics have addressed initial limitations by developing strategies to profile cysteine reactivity within specific subcellular organelles in their native contexts [75]. These strategies include:
- Organelle Enrichment: Fractionation techniques to isolate specific organelles (e.g., mitochondria, endoplasmic reticulum) prior to chemoproteomic analysis, preserving their native pH and redox conditions.
- Organelle-Targeted Reactivity Probes: Designing reactivity-based probes that localize to specific subcellular compartments, enabling in situ labeling of reactive cysteines within living cells.
- Redox-Sensitive Reporters: Using organelle-targeted, redox-sensitive fluorescent proteins (e.g., roGFP) to measure compartment-specific reduction potentials, and pH-sensitive GFP to measure organellar pH [75].
Cysteine-Mediated Redox Signaling in Aging and Disease
Redox Regulation of Aging
Reactive oxygen species (ROS) and hydrogen sulfide (Hâ‚‚S) function as signaling molecules at physiological levels, primarily through the reversible oxidation of reactive protein cysteines [40]. This cysteine-mediated redox signaling plays a crucial role in regulating organismal aging and lifespan. Quantitative analyses of reversible cysteine oxidation across ten mouse tissues have revealed numerous tissue-specific and age-dependent changes in the cysteine redox proteome [40].
Mildly increased levels of ROS or H₂S can extend lifespan in model organisms, and anti-aging interventions—including dietary restriction, reduced insulin/IGF-1 signaling, and mTOR suppression—significantly alter the cysteine redoxome [40]. Specific cysteine oxidation events directly influence longevity; for example, oxidation of a redox-sensitive cysteine in the LET-60 GTPase is required for ROS-mediated lifespan extension in nematodes [40].
Cysteine Oxidative Modifications in Signaling
Cysteine residues undergo a spectrum of oxidative post-translational modifications that regulate protein function:
The enzyme Sulfiredoxin-1 (SRXN1) plays a critical role in reversing cysteine sulfinylation, particularly on peroxiredoxins, thereby restoring their antioxidant activity and maintaining cellular redox homeostasis [21]. SRXN1 expression is transcriptionally regulated by Nrf2 and AP-1, linking its activity to cellular stress response pathways [21]. Beyond its classical function, SRXN1 modulates redox-sensitive signaling pathways governing inflammation, apoptosis, and cell survival, making it a potential therapeutic target in diseases where oxidative stress exacerbates pathology, such as liver disease [21].
The Scientist's Toolkit: Key Research Reagents and Solutions
Table 3: Essential Reagents for Cysteine Reactivity Profiling
| Reagent / Tool | Function / Purpose | Key Features / Examples |
|---|---|---|
| Iodoacetamide (IA) Probe | Electrophilic cysteine labeling | Alkyne handle for bioorthogonal chemistry; e.g., Iodoacetamide-alkyne used in isoTOP-ABPP [76] |
| Isotopic Tags (isoTOP) | Quantitative mass spectrometry | TEV protease sequence, biotin affinity handle, isotopically labeled valine (light/heavy) [76] |
| Organelle-Specific Probes | Targeting subcellular compartments | Designed to localize to mitochondria, ER, etc.; preserves native organelle environment during labeling [75] |
| Redox-Sensitive Fluorophores | Measuring organelle redox potential | roGFP (redox-sensitive GFP) targeted to specific organelles [75] |
| pH-Sensitive Reporters | Measuring organelle pH | pH-sensitive GFP variants [75] |
| Sulfiredoxin-1 (SRXN1) Modulators | Investigating sulfinic acid reduction | Tools to activate or inhibit SRXN1; e.g., Nrf2 inducers to increase SRXN1 expression [21] |
| IA-Linked Chemoproteomic Platforms | Proteome-wide reactivity profiling | Methods like isoTOP-ABPP for quantitative analysis of native cysteine reactivity [76] [78] |
| TB5 | TB5, MF:C15H14BrNOS, MW:336.2 g/mol | Chemical Reagent |
| ALK protein ligand-1 | ALK protein ligand-1, MF:C24H29ClN6O3S, MW:517.0 g/mol | Chemical Reagent |
Cysteine reactivity is precisely tuned through the integrated effects of local electrostatic microenvironments and subcellular localization. Positive charges enhance nucleophilicity by stabilizing the thiolate anion, while organelle-specific pH and redox potentials create specialized landscapes that support compartment-specific cysteine functions. Quantitative chemoproteomic methods such as isoTOP-ABPP have revealed that hyperreactivity is a strong predictor of cysteine functionality, enabling systematic identification of catalytic residues, regulatory sites, and targets of oxidative modification. As our understanding of the nuanced regulation of cysteine reactivity deepens, particularly through advanced databases like CysDB [78], so does our ability to target these residues for therapeutic intervention in redox-related diseases and aging.
Redox regulation, centered on the post-translational modification of protein cysteine residues, has evolved from a concept of oxidative damage to a sophisticated signaling mechanism. A critical advancement in this field is the recognition that reactive oxygen species (ROS) do not signal globally but elicit highly specific effects within distinct subcellular compartments [79]. This compartmentalization, particularly within organelles and endosomes, allows cells to harness redox chemistry for precise physiological regulation, influencing processes from metabolism and cell signaling to protein degradation [80] [81]. The spatial organization of ROS generation, coupled with compartment-specific antioxidant systems and protein scaffolds, creates unique signaling microdomains. This whitepaper delves into the mechanisms, experimental approaches, and therapeutic implications of compartmentalized redox signaling, framing it within the broader context of cysteine residue research.
Fundamental Mechanisms of Compartmentalized Redox Control
The Chemical Versatility of Cysteine Residues
The redox activity of cysteine stems from the electronic structure of its thiol group, which allows multiple oxidation states and a range of reversible modifications, including S-sulfenylation (SOH), S-glutathionylation (SSG), and disulfide bond formation (S-S) [81] [47]. The local protein microenvironment, influencing the thiol's acid dissociation constant (pKa), and the specific subcellular compartment's pH and redox environment collectively determine a cysteine's nucleophilicity and ultimate redox sensitivity [81].
The Role of Membrane Contact Sites (MCSs)
Organelles do not function in isolation but communicate through Membrane Contact Sites (MCSs)—stable, yet dynamic, close appositions between organelle membranes that facilitate the exchange of metabolites and information [82]. For instance, the Endoplasmic Reticulum (ER) and mitochondria connect at structures called Mitochondria-Associated Membranes (MAMs). MAMs are hubs for regulating calcium ion (Ca²âº) transport and lipid exchange, and they host protein complexes like the IP3R-GRP75-VDAC1 complex, which directly facilitates Ca²⺠transfer from the ER to mitochondria [82]. These MCSs are fundamental for coordinating organellar functions and maintaining cellular homeostasis.
Table 1: Key Redox Modifications of Protein Cysteine Residues
| Modification Type | Chemical Formula | Reversibility | Primary Regulatory Role | Example Enzymes/Proteins |
|---|---|---|---|---|
| S-sulfenylation | S-OH | Reversible | Initial signaling oxidation, regulates kinase/phosphatase activity | EGFR, PTP1B |
| Disulfide Bond | S-S | Reversible | Alters protein structure/function; allosteric control | BRSK1, BRSK2 |
| S-glutathionylation | S-SG | Reversible | Protection from over-oxidation; regulates activity | PTP1B, Akt2 |
| S-nitrosylation | S-NO | Reversible | Cross-talk with NO signaling; regulates Ca²⺠channels | |
| Sulfinic Acid | SOâ‚‚H | Difficult to reverse | Can be regulatory (e.g., PRDXs) | Peroxiredoxins |
| Sulfonic Acid | SO₃H | Irreversible | Often associated with oxidative damage |
Redox Signaling in Specific Subcellular Compartments
Endosomal Compartments
Endosomes are a prime example of a specialized redox signaling compartment. Following ligand stimulation, Receptor Tyrosine Kinases (RTKs) like the Epidermal Growth Factor Receptor (EGFR) are internalized into endosomes [81]. The endosomal membrane can host NADPH oxidase 4 (NOX4), which generates superoxide and subsequently hydrogen peroxide (Hâ‚‚Oâ‚‚) locally [79]. This creates a confined redox-active environment where Hâ‚‚Oâ‚‚ can oxidize specific cysteine residues on signaling proteins. For example, local oxidation and inactivation of the phosphatase PTP1B within the endosome prevents the dephosphorylation of EGFR, thereby promoting the receptor's recycling back to the plasma membrane and sustaining its signaling activity [81]. This mechanism demonstrates how compartmentalization enables spatial control of signaling amplitude and duration.
The Mitochondria and MAMs
Mitochondria are major sites of ROS generation, with complexes I and III of the electron transport chain being primary sources of superoxide [79]. The redox state within the mitochondrial matrix is tightly regulated by local antioxidants like Peroxiredoxin 3 (Prx3) and Superoxide Dismutase 2 (SOD2) [79]. At the MAMs, redox signaling is intricately linked with calcium homeostasis. The ER releases Ca²⺠through the inositol 1,4,5-trisphosphate receptor (IP3R), which is taken up by mitochondria via the voltage-dependent anion channel (VDAC1), a process facilitated by the chaperone GRP75 [82]. This Ca²⺠flux is crucial for regulating mitochondrial metabolism and can, in turn, influence mitochondrial ROS production. Furthermore, the ER-mitochondria interface is critical for lipid transfer, such as the conversion of phosphatidylserine (PS) to phosphatidylethanolamine (PE) [82].
The Cytosol and Plasma Membrane
Cytosolic ROS signaling is often initiated by NADPH oxidases at the plasma membrane, such as NOX2 [79]. In skeletal muscle, NOX2 is located in the transverse tubules, and its activation during exercise produces superoxide. After conversion to Hâ‚‚Oâ‚‚, it can enter the cytosol to regulate adaptive signaling pathways [79]. The cytosolic redox environment is buffered by systems like the Thioredoxin (Trx) and Glutathione (GSH) systems, which also play a role in reducing oxidative modifications on proteins, thus terminating redox signals [47].
Figure 1: Compartmentalized Redox Signaling in EGFR Endosomal Trafficking. Local ROS production from NOX4 within the endosome oxidizes and inactivates PTP1B, preventing EGFR dephosphorylation and promoting receptor recycling and sustained signaling [81].
Experimental Approaches and Research Tools
Genetically Encoded Redox Biosensors
A pivotal technological advancement for studying compartmentalized redox signaling is the development of compartment-targeted, genetically encoded biosensors [79]. For instance, the Hâ‚‚Oâ‚‚-specific biosensor Orp1-roGFP can be targeted to the mitochondrial matrix to monitor real-time changes in Hâ‚‚Oâ‚‚ levels within that specific compartment. A key finding using this tool was that mitochondrial Hâ‚‚Oâ‚‚ levels in skeletal muscle actually decrease during moderate-intensity exercise, challenging the simple mitohormesis model and highlighting the compartment-specific nature of ROS dynamics [79].
Structural and Proteomic Techniques
Understanding the structural context of redox-active cysteines is essential. Techniques such as X-ray crystallography and molecular modeling have revealed that cysteine residues in kinases often reside in functionally critical regions, such as the activation loop or near the ATP-binding site [83] [84]. Proteome-wide analyses, like those in the Oximouse database, help quantify cysteine oxidation in vivo and correlate it with local structural environments, revealing that non-conserved, solvent-exposed cysteines are often the most reactive [83].
Table 2: The Scientist's Toolkit: Key Reagents and Methods for Compartmentalized Redox Research
| Tool/Method | Specific Example | Function/Application | Key Insight Enabled |
|---|---|---|---|
| Genetically Encoded Biosensors | Orp1-roGFP (targeted to mitochondria/cytosol) | Real-time measurement of Hâ‚‚Oâ‚‚ in specific compartments | Mitochondrial Hâ‚‚Oâ‚‚ decreases during muscle contraction [79] |
| Redox Proteomics | Oximouse database, ICAT, OxMRM | Quantifies oxidative modifications on specific cysteine residues proteome-wide | Identifies cysteines in non-conserved, solvent-exposed regions as highly reactive [83] |
| Structural Biology | X-ray Crystallography, Molecular Modelling | Visualizes disulfide bonds and local protein environment of cysteines | Revealed disulfide bonds in BRSK1/BRSK2 kinases cause allosteric inactivation [84] |
| Covalent Inhibitors | Afatinib (EGFR inhibitor) | Irreversibly binds to cysteine residues in kinase active sites | Validates specific cysteines as druggable targets; therapeutic strategy [84] |
| MCS Manipulation | siRNA against MCS tethering proteins (e.g., VAP, MFN2) | Disrupts specific organelle interactions to study functional outcomes | Established MAMs as hubs for Ca²⺠transfer and lipid synthesis [82] |
Detailed Experimental Protocol: Assessing Compartment-Specific Hâ‚‚Oâ‚‚ Dynamics During Endosomal Signaling
This protocol outlines a combined approach using biosensor transfection and ligand stimulation to monitor redox changes in endosomes.
- Primary Objective: To quantify changes in Hâ‚‚Oâ‚‚ levels within endosomal compartments during EGFR signaling.
- Key Reagents:
- Expression Constructs: Plasmid DNA for endosome-targeted roGFP2-Orp1 (e.g., fused to Rab5a for early endosomes).
- Cell Line: HEK293 or HeLa cells.
- Stimulus: Recombinant human EGF.
- Inhibitors: Diphenyleneiodonium (DPI, NOX inhibitor) or VAS2870 (specific NOX inhibitor).
- Imaging Buffer: Hanks' Balanced Salt Solution (HBSS) with low autofluorescence.
- Methodology:
- Cell Transfection: Transfect cells with the endosome-targeted roGFP2-Orp1 construct using a standard method (e.g., lipofection) 24-48 hours prior to imaging.
- Image Acquisition: Plate transfected cells on glass-bottom dishes. Use a confocal or fluorescence microscope capable of sequential imaging at two excitation wavelengths (e.g., 405 nm and 488 nm) for ratiometric analysis. Set environmental control to 37°C and 5% CO₂.
- Experimental Treatment:
- Acquire a 2-minute baseline ratiometric measurement (405/488 nm excitation, 510 nm emission).
- Add EGF (e.g., 100 ng/mL) to the imaging buffer and continue ratiometric acquisition for 15-30 minutes.
- For inhibitor controls, pre-treat cells with a NOX inhibitor (e.g., 10 µM VAS2870) for 30-60 minutes before EGF stimulation.
- Data Analysis:
- Calculate the 405/488 nm emission ratio for each time point.
- Normalize the ratio to the baseline (pre-stimulation) value, expressing data as Fold Change over Baseline.
- Compare the peak ratio and kinetics between EGF-stimulated and inhibitor-treated groups using appropriate statistical tests (e.g., t-test, ANOVA).
- Expected Outcome: EGF stimulation should cause a rapid, transient increase in the roGFP2-Orp1 oxidation ratio specifically within endosomal compartments, which is attenuated by NOX inhibition, indicating NOX-derived Hâ‚‚Oâ‚‚ production in this specific locale [81] [79].
Functional Consequences and Therapeutic Implications
Regulation of Kinase Activity and Signaling Pathways
Redox modification directly regulates the activity of key protein kinases. Research on the brain-selective kinases BRSK1 and BRSK2 revealed that oxidation of specific cysteine residues leads to the formation of disulfide bonds, which allosterically repress kinase activity [84]. Conversely, oxidation of a specific cysteine (Cys797) in EGFR can enhance its catalytic activity [81] [84]. It is estimated that roughly 10% of human protein kinases may be subject to redox regulation, highlighting the breadth of this control mechanism [84]. The cross-talk between phosphorylation and cysteine oxidation adds a complex layer of regulation to cellular signaling networks.
Implications for Drug Discovery: Covalent Inhibitors
The presence of redox-active cysteine residues in functionally important regions of proteins, particularly kinases, presents a unique therapeutic opportunity. Covalent inhibitors are small molecules designed to permanently bind to these cysteine residues, blocking protein activity [84]. This strategy has been successfully employed for kinases like EGFR, with drugs like afatinib and osimertinib receiving FDA approval. The systematic mapping of the kinome "cysteinome" continues to reveal new, druggable cysteine residues, paving the way for a new generation of targeted therapies that exploit the native redox biochemistry of their targets [83] [84].
Figure 2: Workflow for Developing Covalent Inhibitors Targeting Redox-Active Cysteines. The process integrates proteomic data, structural biology, and functional assays to design drugs that irreversibly bind to specific cysteine residues, offering a potent therapeutic strategy [83] [84].
The study of compartmentalized redox signaling represents a frontier in understanding cellular regulation. The precise spatiotemporal control of redox signals, mediated through the chemical modification of specific cysteine residues in organelles and endosomes, is fundamental to physiology and disease. Future research, powered by advanced biosensors, structural techniques, and proteomics, will continue to decode this complex language. This deeper understanding promises not only to elucidate fundamental biological processes but also to unlock novel, precise therapeutic strategies for a wide range of diseases, from cancer to neurodegenerative disorders, by targeting the very redox switches that govern cellular life.
The "Antioxidant Paradox" describes the stark contrast between the demonstrated role of oxidative stress in chronic diseases and the consistent failure of broad-spectrum antioxidant supplements in clinical trials. This whitepaper reframes this paradox through the lens of modern redox biology, emphasizing that oxidative signaling occurs through specific, reversible post-translational modifications of protein cysteine residues rather than through random macromolecular damage. We explore how non-targeted antioxidant approaches disrupt crucial redox signaling pathways, while emerging targeted strategies—including sulfur-based redox modulators, protein kinase-specific regulators, and enzyme-specific antioxidants—offer promising alternatives by working within physiological redox architecture. By integrating recent advances in cysteine redox proteomics, structural biology, and targeted drug delivery, this review provides a framework for developing next-generation redox therapeutics that respect the spatial and temporal specificity of native redox signaling networks.
The field of redox biology has undergone a fundamental paradigm shift. Reactive oxygen species (ROS), once exclusively considered damaging molecules, are now recognized as essential signaling mediators that regulate a vast array of cellular processes through specific, reversible modifications of cysteine residues in proteins [40] [85]. This dual nature of ROS creates a fundamental challenge for therapeutic intervention: while excessive ROS production contributes to pathology, global ROS scavenging disrupts essential redox signaling.
The antioxidant paradox emerges from this complexity. Broad-spectrum antioxidants typically function as indiscriminate electron donors that neutralize various ROS without specificity for their source, concentration, or physiological function [86] [87]. This approach fails to distinguish between pathological oxidative damage and physiological redox signaling, leading to the disruption of essential cellular processes. In contrast, targeted approaches aim to modulate specific redox nodes within cellular signaling networks, particularly those involving cysteine residues that have evolved as specialized redox sensors [40] [84].
At the core of this paradox is the realization that cysteine residues are not merely passive targets of oxidative damage but sophisticated regulatory elements. The human proteome contains approximately 210,000 cysteine residues, with thousands exhibiting sensitivity to oxidants [40]. These reactive cysteines undergo a complex repertoire of post-translational modifications including sulfenylation (-SOH), disulfide bond formation (-S-S-), glutathionylation (-SSG), and persulfidation (-SSH), each with distinct functional consequences and reversibility profiles [21] [40]. The specificity of redox signaling is further enhanced by compartmentalization, with precise subcellular localization of ROS production and antioxidant defense systems creating microdomains of redox regulation [40] [88].
Molecular Mechanisms of Cysteine Redox Signaling
The Cysteine Redox Code
Cysteine residues serve as fundamental information processing nodes in cellular signaling networks due to the unique redox properties of their thiol groups. The reactivity of a specific cysteine is determined by its molecular environment, with key factors including:
- Local protein microenvironment - Adjacent basic amino acids can stabilize the thiolate anion (Sâ»), increasing reactivity by up to 1000-fold
- Solvent accessibility - Surface-exposed cysteines demonstrate different reactivity profiles compared to buried residues
- pKa modulation - Hydrogen bonding networks and electrostatic effects can significantly lower thiol pKa values
- Metal coordination - Cysteine residues frequently participate in zinc finger domains and iron-sulfur clusters [21]
The hierarchy of cysteine oxidative modifications follows a well-defined chemical pathway, with each modification exhibiting distinct stability and functional consequences:
Table 1: Hierarchy of Cysteine Oxidative Modifications
| Modification | Chemical Formula | Reversibility | Primary Reductase | Signaling Role |
|---|---|---|---|---|
| Sulfenylation | -SOH | Fully reversible | Thioredoxin/Glutaredoxin | Early signaling event |
| Disulfide bond | -S-S- | Fully reversible | Thioredoxin/Glutaredoxin | Structural regulation |
| Glutathionylation | -SSG | Fully reversible | Glutaredoxin | Metabolic regulation |
| Persulfidation | -SSH | Fully reversible | Thioredoxin | Hâ‚‚S-mediated signaling |
| Sulfinylation | -SOâ‚‚H | Reversible | Sulfiredoxin | Sustained activation |
| Sulfonylation | -SO₃H | Irreversible | None | Terminal inactivation |
This "redox code" enables precise temporal and spatial control over protein function, with different modifications encoding different biological information [21] [40].
Compartmentalized ROS Production and Antioxidant Defense
Cellular redox signaling depends on tightly controlled, compartmentalized ROS production. Major sources include:
- Mitochondrial electron transport chain - Complex I and III leak electrons to oxygen, forming superoxide (O₂•â») as a byproduct of oxidative phosphorylation [88]
- NADPH oxidases (NOX family) - Transmembrane enzymes dedicated to regulated ROS production for signaling purposes [40] [88]
- Endoplasmic reticulum - Oxidative protein folding generates Hâ‚‚Oâ‚‚ as a byproduct through ERO1 and QSOX activity [40]
- Peroxisomes - Contain multiple oxidases that produce Hâ‚‚Oâ‚‚ during metabolic reactions [40]
Complementing these production sites, a sophisticated antioxidant defense system provides compartment-specific protection:
- Superoxide dismutases (SOD1/2/3) catalyze the conversion of O₂•⻠to H₂O₂
- Catalase decomposes Hâ‚‚Oâ‚‚ to water and oxygen in peroxisomes
- Glutathione peroxidases (GPX) reduce Hâ‚‚Oâ‚‚ and lipid hydroperoxides using glutathione
- Peroxiredoxins (Prx) catalyze the reduction of Hâ‚‚Oâ‚‚, organic hydroperoxides, and peroxynitrite [40] [85]
The regeneration of antioxidant systems is particularly crucial. The thioredoxin (Trx) and glutathione (GSH) systems maintain redox homeostasis by reducing oxidized proteins, while sulfiredoxin (SRXN1) plays a specialized role in reducing hyperoxidized peroxiredoxins, restoring their peroxidase activity through an ATP-dependent mechanism [21].
Figure 1: Cysteine-Mediated Redox Signaling Network
Why Broad-Spectrum Antioxidants Fail: A Mechanistic Analysis
Disruption of Physiological Redox Signaling
Broad-spectrum antioxidants, including vitamin C, vitamin E, and various plant polyphenols, typically function as non-specific redox-active compounds that scavenge multiple ROS types without molecular specificity [86] [87]. While effective at reducing global oxidative damage markers in vitro, these compounds consistently fail in clinical trials for chronic diseases because they disrupt essential redox signaling circuits:
- Interference with metabolic regulation - ROS serve as essential second messengers in insulin signaling, with Hâ‚‚Oâ‚‚ transiently inactivating protein tyrosine phosphatases to enhance insulin receptor activation [85]
- Impairment of immune function - Phagocytic cells require ROS production for microbial killing and inflammatory signaling; global antioxidant administration can compromise host defense [85] [88]
- Disruption of hypoxic adaptation - Hypoxia-inducible factor (HIF) stability is regulated by ROS-sensitive prolyl hydroxylases; antioxidants can interfere with proper hypoxic signaling [88]
- Alteration of kinase pathways - Multiple protein kinases, including EGFR, Src, and BRSK1/2, are regulated through reversible cysteine oxidation; non-specific antioxidants disrupt this regulation [84]
The failure of broad-spectrum antioxidants is particularly evident in large-scale human trials. The SELECT trial (Selenium and Vitamin E Cancer Prevention Trial) found not only no benefit but increased risks of certain conditions, illustrating the potential dangers of disrupting redox homeostasis [85].
Pharmacokinetic and Chemical Limitations
Beyond their biological non-specificity, broad-spectrum antioxidants face significant pharmacokinetic and chemical challenges:
Table 2: Limitations of Broad-Spectrum Antioxidants
| Limitation | Impact on Efficacy | Examples |
|---|---|---|
| Poor bioavailability | Limited tissue distribution and cellular uptake | Many polyphenols have <5% bioavailability |
| Non-specific distribution | Inability to reach relevant subcellular compartments | Vitamin C accumulates in cytosol but not mitochondria |
| Pro-oxidant effects | Can autoxidize to generate ROS under certain conditions | Vitamin C in the presence of transition metals |
| Rapid metabolism | Short half-life prevents sustained therapeutic effect | Rapid conjugation and excretion of flavonoid compounds |
| Lack of concentration-dependent effects | Biphasic responses with opposite effects at different doses | Low doses may enhance ROS signaling while high doses suppress it |
The chemical mechanisms of traditional antioxidants further limit their specificity. Most operate through single-electron transfer (SET) or hydrogen atom transfer (HAT) mechanisms that lack target specificity [86] [87]. These fundamental limitations have prompted the development of targeted approaches that work within, rather than against, physiological redox signaling architecture.
Targeted Redox-Based Therapeutic Approaches
Protein Kinase Redox Regulation
Recent structural biology advances have revealed that approximately 10% of human protein kinases contain redox-sensitive cysteine residues in critical functional regions, representing a novel targeting strategy [84]. The kinase cysteinome comprises spatially clustered cysteine residues that regulate catalytic activity through reversible disulfide bond formation:
- BRSK1 and BRSK2 - Oxidation of specific cysteine pairs forms disulfide bonds that repress kinase activity by stabilizing inactive conformations [84]
- EGFR - Cysteine oxidation in the activation loop enhances kinase activity and promotes dimerization [84]
- c-Src and LRRK2 - Contain cysteine residues whose oxidation modulates catalytic function through distinct mechanisms [84]
These regulatory cysteine residues create opportunities for covalent kinase inhibitors that specifically target redox-sensitive cysteines rather than the conserved ATP-binding pocket. Over forty FDA-approved drugs utilize this mechanism, primarily targeting cysteine residues in the front region (F2 position) of the kinase active site [84].
Figure 2: Redox Regulation of Protein Kinases
Nrf2-SRXN1 Redox Axis Targeting
The transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) represents a master regulator of the cellular antioxidant response, controlling the expression of over 230 genes involved in detoxification and redox homeostasis [21]. Unlike broad-spectrum antioxidants, Nrf2 activators enhance native cellular defense systems in a coordinated manner:
- SRXN1 regulation - SRXN1 has been identified as a direct Nrf2 target gene, with a conserved antioxidant response element (ARE) in its promoter region [21]
- Context-specific activity - Nrf2 activation induces a coordinated transcriptional program adapted to cell type and stress status
- Feedforward regulation - SRXN1 enhances peroxiredoxin recycling, creating a positive feedback loop that boosts Hâ‚‚Oâ‚‚ scavenging capacity [21]
The Nrf2-SRXN1 axis demonstrates the principle of systems-level redox intervention that enhances native antioxidant capacity without disrupting specific signaling functions of ROS. Small molecule Nrf2 activators such as sulforaphane (from broccoli sprouts) and synthetic triterpenoids have shown promising results in preclinical models of neurodegenerative and metabolic diseases [21].
Mitochondria-Targeted Antioxidants
Mitochondria represent both a primary source and target of ROS, making them critical sites for redox damage in aging and neurodegeneration. Mitochondria-targeted antioxidants address the compartmentalization issue by selectively accumulating in the mitochondrial matrix:
Table 3: Mitochondria-Targeted Antioxidants
| Compound | Mechanism | Therapeutic Potential |
|---|---|---|
| MitoQ | Ubiquinone linked to TPP⺠cation accumulates in mitochondria, reduces local ROS | Neurodegenerative diseases, metabolic disorders |
| MitoVitE | Vitamin E conjugated to TPP⺠protects mitochondrial membranes from peroxidation | Ischemia-reperfusion injury, aging |
| MitoApocynin | TPPâº-conjugated apocynin derivative inhibits mitochondrial ROS production | Parkinson's disease models |
| SS-31 (Elamipretide) | Aromatic-cationic peptide that binds to cardiolipin in inner mitochondrial membrane | Mitochondrial myopathies, neurodegenerative diseases |
These targeted compounds demonstrate significantly enhanced efficacy compared to their non-targeted analogs in preclinical models, validating the importance of subcellular targeting in redox-based therapeutics [89].
Experimental Approaches for Targeted Redox Research
Redox Proteomics and Cysteine Chemoproteomics
Comprehensive analysis of the cysteine redox proteome requires specialized methodologies that capture the labile nature of oxidative modifications:
Protocol 1: Quantitative Redox Proteomics Using Iodoacetyl Tandem Mass Tags (iodoTMT)
- Cell lysis under controlled redox conditions - Include alkylating agents to prevent artificial oxidation during extraction
- Free thiol blocking - Treat with N-ethylmaleimide (NEM) to block reduced cysteine residues
- Selective reduction of oxidized species - Use ascorbate for sulfenic acids or specific reductases for disulfides
- Tagging with isobaric labels - Treat with iodoacetyl TMT reagents to label newly reduced cysteines
- Multiplexed LC-MS/MS analysis - Quantify relative oxidation states across multiple conditions simultaneously
This approach has revealed tissue- and age-dependent changes in the cysteine redox proteome, identifying thousands of oxidant-sensitive cysteines with functional roles in diverse biological processes [40].
Structural Biology of Redox-Sensitive Proteins
High-resolution structural analysis is essential for understanding how cysteine oxidation regulates protein function:
Protocol 2: X-Ray Crystallography of Redox-Sensitive Proteins
- Site-directed mutagenesis - Replace redox-sensitive cysteines with serine or alanine to stabilize specific oxidation states
- Crystallization under anaerobic conditions - Maintain controlled redox environment during crystal formation
- Post-crystallization oxidation - Soak crystals in oxidizing agents to generate specific cysteine modifications
- High-resolution data collection - Resolve electron density for sulfur atoms and oxidative modifications
- Molecular dynamics simulations - Model the functional consequences of oxidation-induced conformational changes
This approach has revealed how disulfide bond formation in BRSK1 and BRSK2 stabilizes specific autoinhibitory conformations, providing a structural basis for redox regulation of kinase activity [84].
In Vivo Redox Biosensing
Real-time monitoring of redox dynamics in living systems requires genetically encoded biosensors:
Protocol 3: Monitoring Subcellular Hâ‚‚Oâ‚‚ Dynamics with HyPer Biosensor
- Sensor expression - Transfect with HyPer (a Hâ‚‚Oâ‚‚-sensitive fluorescent protein) targeted to specific subcellular compartments
- Ratiometric imaging - Measure excitation ratio 420/490 nm (emission 516 nm) to quantify Hâ‚‚Oâ‚‚ concentration
- Calibration - Treat with bolus Hâ‚‚Oâ‚‚ and dithiothreitol to establish minimum and maximum ratio values
- Time-lapse imaging - Monitor Hâ‚‚Oâ‚‚ dynamics in response to physiological stimuli or pharmacological interventions
- Data analysis - Convert ratio values to Hâ‚‚Oâ‚‚ concentrations using established calibration curves
This approach has revealed spatially restricted Hâ‚‚Oâ‚‚ gradients that function in specific signaling pathways, highlighting the compartmentalized nature of redox signaling [40].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for Targeted Redox Research
| Reagent Category | Specific Examples | Research Application | Key Function |
|---|---|---|---|
| Cysteine-directed probes | IodoTMT, BIAM, NEM-biotin | Redox proteomics | Selective labeling of redox-sensitive cysteines |
| Genetically encoded biosensors | HyPer, roGFP, Grx1-roGFP | Live-cell imaging | Real-time monitoring of subcellular Hâ‚‚Oâ‚‚ and glutathione redox potential |
| Targeted antioxidant compounds | MitoQ, MitoTEMPO, MitoSNO | Mitochondrial redox studies | Selective scavenging of mitochondrial ROS |
| Nrf2 pathway modulators | Sulforaphane, Bardoxolone methyl, ML385 | Antioxidant response studies | Activation or inhibition of Nrf2-mediated transcription |
| SRXN1 activity tools | Recombinant SRXN1, Anti-SRXN1 antibodies, SRXN1 inhibitors | Sulfinic acid reduction studies | Investigation of peroxiredoxin recycling pathways |
| Cysteine oxidation-specific antibodies | Anti-sulfenylate (DCP-Rho1), Anti-nitrotyrosine | Immunodetection of oxidative modifications | Detection and localization of specific cysteine oxidative modifications |
| Hâ‚‚Oâ‚‚ generators | Glucose oxidase, AAPH, PLVA-PEG | Controlled oxidative stress models | Generation of specific ROS under controlled conditions |
| Licarbazepine-d8 | Licarbazepine-d8, MF:C15H14N2O2, MW:262.33 g/mol | Chemical Reagent | Bench Chemicals |
| DJ101 | DJ101, MF:C23H20N4O3, MW:400.4 g/mol | Chemical Reagent | Bench Chemicals |
The resolution to the antioxidant paradox lies in embracing the complexity of cellular redox regulation rather than attempting to override it with simplistic scavenging approaches. Targeted redox therapeutics represent a paradigm shift from global antioxidant suppression to precise modulation of specific redox nodes within cellular signaling networks. Future advances in this field will depend on:
- Spatiotemporal precision - Developing approaches that target specific subcellular compartments and timeframes of redox signaling
- Redox proteome mapping - Comprehensive characterization of context-dependent cysteine oxidation across tissues, cell types, and disease states
- Structural redox biology - High-resolution understanding of how oxidative modifications alter protein function at atomic detail
- Personalized redox medicine - Tailoring interventions based on individual variations in redox regulation and antioxidant capacity
The continued evolution from broad-spectrum antioxidants to targeted redox modulators promises to finally deliver on the therapeutic potential of redox-based interventions while respecting the sophisticated signaling functions of reactive oxygen species in physiological regulation.
Redox regulation, particularly the oxidative post-translational modifications (oxiPTMs) of protein cysteine residues, represents a fundamental signaling mechanism that transcends organismal boundaries. These modifications act as molecular switches that dynamically regulate protein function, signaling pathways, and cellular adaptation to environmental stress [46] [47]. The cysteine thiol group (–SH) is highly reactive toward various reactive oxygen and nitrogen species, enabling the formation of reversible modifications including S-sulfenylation (–SOH), S-nitrosylation (–SNO), S-glutathionylation (–SSG), and persulfidation (–SSH) [46] [90]. Under physiological conditions, cells maintain redox homeostasis through sophisticated antioxidant systems, but disruption of this equilibrium is intimately linked to disease pathogenesis across multiple organ systems [47] [90].
This technical guide provides a structured framework for designing rigorous experimental approaches in redox biology, focusing specifically on the validation of cysteine-mediated redox regulation from initial discovery in model systems through to establishing human disease relevance. We synthesize current methodologies, computational tools, and validation strategies to enable robust translational research in this rapidly evolving field.
Model Organism Selection for Redox Studies
Selecting appropriate model organisms is paramount for establishing a foundation for translational redox research. Each model offers distinct advantages for specific research questions, and the choice should align with the biological context and practical constraints of the study.
Table 1: Model Organisms in Redox Research with Key Applications
| Organism | Key Advantages | Redox-Related Applications | Technical Limitations |
|---|---|---|---|
| Arabidopsis thaliana | Well-characterized redox signaling networks; genetic tractability; ideal for studying photosynthetic redox regulation | Photosynthetic electron transport; environmental stress responses; redox signaling in development [46] [91] | Limited direct translational relevance to mammalian systems |
| Mammalian models (Mouse, Rat) | Conserved redox systems; genetically engineered strains; physiological relevance to human disease | NRF2-Keap1 pathway function; cardiovascular redox pathophysiology; neurological disease mechanisms [92] [93] [94] | Higher maintenance costs; ethical considerations; complex genetic manipulation |
| Saccharomyces cerevisiae | Rapid generation time; powerful genetic tools; well-characterized redox relay mechanisms | Fundamental redox signaling mechanisms; conserved thiol-based regulatory pathways [46] | Limited tissue complexity; divergent from mammalian physiology in specific aspects |
Beyond these established models, emerging evidence suggests that comparative approaches across multiple species can yield powerful insights into conserved versus specialized redox mechanisms. For studies focusing on central nervous system pathologies, the mouse UPOAO (unilateral pterygopalatine ophthalmic artery occlusion) model has demonstrated particular utility for investigating retinal ischemia-reperfusion injury and testing potential redox-modulating therapeutics [94].
Experimental Workflows for Redox Proteomic Investigation
Comprehensive analysis of cysteine redox modifications requires specialized workflows that capture their labile and dynamic nature. The following section outlines two complementary approaches that enable proteome-wide profiling of cysteine oxidation states.
CysQuant Workflow for Absolute Oxidation Quantification
The CysQuant methodology enables simultaneous quantification of cysteine oxidation degrees and protein abundances using light/heavy iodoacetamide isotopologues for differential labeling of reduced and reversibly oxidized cysteines [91].
Table 2: Key Steps in the CysQuant Experimental Protocol
| Step | Procedure | Critical Parameters | Purpose |
|---|---|---|---|
| 1. Sample Preparation | Grind tissue in liquid nitrogen; transfer to trichloroacetic acid (TCA) buffer; precipitate proteins | Rapid processing to preserve native redox states; acidic conditions to protonate thiols | Stabilize in vivo oxidation states before artificial oxidation occurs |
| 2. Block Reduced Thiols | Label reduced cysteine thiols with light iodoacetamide (IAM0) in denaturing buffer | Strong denaturation to expose all reduced thiols; complete alkylation to prevent post-lysis oxidation | Permanently tag initially reduced cysteine populations |
| 3. Reduce Oxidized Thiols | Capture proteins on S-Trap column; reduce with TCEP | Efficient trapping to prevent sample loss; strong reducing agent to revert reversible oxidations | Convert reversibly oxidized thiols to reduced form for labeling |
| 4. Label Newly Reduced Thiols | Alkylate with heavy iodoacetamide (IAM4) | Differential mass tag for MS distinction; complete labeling of all newly reduced thiols | Tag the originally oxidized cysteine population |
| 5. MS Analysis | Trypsin digestion; LC-MS/MS analysis via DDA or DIA modes | Use of plexDIA with in silico predicted spectral libraries; high-resolution mass spectrometry | Quantify IAM0/IAM4 peptide pairs to calculate oxidation percentage |
This workflow has been successfully applied to quantify an average of 18% cysteine oxidation in Arabidopsis thaliana, including identification of highly oxidized cysteines forming disulfide bridges in AlphaFold2 predicted structures [91]. The method is particularly valuable for assessing redox responses to environmental perturbations, such as the well-characterized increased reduction of Calvin-Benson cycle enzymes in plants exposed to excessive light [91].
SICyLIA Workflow for Comparative Redox Profiling
The Stable Isotope Cysteine Labelling with Iodoacetamide (SICyLIA) approach provides an unbiased method for proteome-wide assessment of cysteine oxidation dynamics without requiring enrichment steps [50].
The SICyLIA method has demonstrated exceptional utility in models of chronic oxidative stress, such as fumarate hydratase-deficient cells, where it identified specific metabolic adaptations through oxidation of distinct metabolic proteins [50]. This approach enables researchers to identify cysteine residues that undergo significant oxidation changes under pathological conditions, providing a foundation for further mechanistic investigation.
The Scientist's Toolkit: Essential Research Reagents and Technologies
Contemporary redox research requires specialized reagents and technologies designed to capture, quantify, and manipulate cysteine oxidation events. The following toolkit summarizes critical resources for conducting rigorous investigations in this field.
Table 3: Essential Research Reagents for Redox Signaling Studies
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| Chemoselective Probes | Dimedone and derivatives (DCP-N3, Dyn-2); nitrobenzene-based warheads | Selective labeling and detection of sulfenic acid modifications; enables enrichment and visualization of specific oxiPTMs [6] |
| Isotopic Labeling Reagents | Light/heavy iodoacetamide (IAM0/IAM4); iodoTMT; OxICAT tags | Differential labeling of reduced vs. oxidized cysteine pools; enables MS-based quantification of oxidation states [46] [91] [50] |
| Enrichment Tools | Biotin-switch assay; resin-assisted capture (RAC); antibody-based purification | Isolation of cysteine-containing peptides or specific oxiPTMs; reduces sample complexity for enhanced detection sensitivity [46] |
| Computational Prediction Tools | CysQuant; BiGRUD-SA; DLF-Sul; Plant PTM Viewer | Prediction of redox-sensitive cysteine residues; functional annotation of redox-modified proteins; integration with structural data [46] |
| Site-Specific Manipulation Tools | Photocaged cysteine sulfoxides (DMNB-caged); genetic code expansion systems | Precise incorporation and activation of SOH modifications at specific sites; enables causal relationship establishment [6] |
This toolkit continues to expand with emerging technologies, particularly in the realm of site-specific manipulation. Recent innovations include the integration of bioorthogonal cleavage chemistry with genetic code expansion for precise incorporation of sulfenic acid modifications in proteins of interest, enabling controlled activation of redox events with temporal precision [6]. Additionally, the development of redox-targeted covalent inhibitors (TCIs) with moderately reactive warheads (e.g., nitroacetamide groups) offers promising approaches for selectively blocking SOH modifications at specific targets [6].
Computational and Analytical Approaches for Data Integration
The complexity of redox signaling networks necessitates sophisticated computational approaches to extract biological insights from large-scale proteomic datasets. Machine learning frameworks have emerged as powerful tools for predicting redox-sensitive residues and characterizing redox-dependent signaling networks [46].
Several specialized algorithms have been developed specifically for redox proteomics applications. The CysQuant computational pipeline integrates with DIA-NN software for spectral library generation and quantification of light/heavy peptide pairs, enabling accurate determination of cysteine oxidation percentages [91]. For prediction of specific modification types, tools such as BiGRUD-SA and DLF-Sul utilize deep learning frameworks to identify cysteine residues susceptible to sulfenic acid formation with high precision [46]. The Plant PTM Viewer provides a specialized resource for contextualizing redox modifications within broader post-translational modification networks in plant systems [46].
Effective data interpretation requires integration of redox proteomic findings with complementary datasets. Correlation with transcriptomic data can distinguish direct redox regulation from transcriptional changes, while structural modeling using AlphaFold2 predictions can rationalize how specific cysteine modifications impact protein function [46] [91]. Additionally, metabolic pathway analysis places redox-sensitive enzymes within their functional contexts, revealing how coordinated oxidation of enzyme families can redirect metabolic flux under stress conditions [50].
Validation Strategies for Establishing Human Disease Relevance
The transition from initial discovery in model systems to establishing pathophysiological relevance in human disease represents a critical challenge in redox research. A multi-tiered validation strategy provides the most compelling evidence for clinical significance.
Biochemical Validation of Causal Mechanisms
Beyond correlation, establishing causal relationships between specific cysteine oxidations and functional consequences requires sophisticated experimental approaches. Site-directed mutagenesis of redox-sensitive cysteines to redox-insensitive residues (e.g., serine) represents a foundational approach, but this strategy may disrupt structural disulfide bonds or metal binding sites [6]. More refined techniques now enable site-specific manipulation of redox events, such as incorporating photocaged cysteine sulfoxides via genetic code expansion to enable light-controlled activation of specific SOH modifications [6].
Targeted covalent inhibitors (TCIs) designed with moderately reactive warheads offer complementary loss-of-function approaches. These compounds can selectively engage specific oxidized cysteine residues, blocking their redox regulation without affecting other cysteine functions [6]. For example, dimedone-based compounds paired with target-binding modules have shown promise for selectively inhibiting SOH-mediated regulation of protein tyrosine phosphatases [6].
Physiological and Pathophysiological Validation
Demonstrating disease relevance requires validation in physiological contexts and patient-derived samples. The following diagram illustrates an integrated validation workflow for establishing the clinical significance of redox modifications identified in model systems.
The application of redox proteomics to patient-derived samples represents a crucial step in validation. Studies have successfully quantified cysteine oxidation patterns in complex human tissues, including kidney biopsies, revealing disease-specific oxidation signatures [50]. Similarly, analysis of circulating proteins in biofluids provides opportunities to identify redox biomarkers that reflect systemic oxidative stress burden or specific pathological processes [90] [50].
For neurological disorders, emerging evidence demonstrates the potential of targeting redox pathways for therapeutic benefit. In central retinal artery occlusion, kynurenine-mediated activation of the AhR-Nrf2 signaling axis has shown neuroprotective effects by mitigating oxidative stress in retinal ganglion cells, highlighting how understanding endogenous redox regulatory mechanisms can inform therapeutic development [94].
The field of cysteine redox regulation continues to evolve with increasingly sophisticated tools for detecting, quantifying, and manipulating specific oxidative modifications. By implementing rigorous experimental designs that traverse from model organisms to human disease validation, researchers can establish robust evidence for the functional significance of redox events in physiological and pathological processes. The integrated frameworks presented in this guide provide a roadmap for advancing our understanding of redox biology with translational impact, potentially informing the development of targeted therapies for conditions characterized by redox dysregulation.
Validating Redox Targets and Comparative Mechanisms in Disease Pathogenesis
Sulfiredoxin-1 (SRXN1) has emerged as a critical regulator of cellular redox homeostasis through its unique ability to reduce cysteine sulfinic acid (Cys-SOâ‚‚H) modifications in target proteins. Recent investigations across various liver disease models, particularly hepatocellular carcinoma (HCC) and liver fibrosis, have validated SRXN1 as a promising therapeutic target. This technical review synthesizes current evidence demonstrating that SRXN1 modulates key signaling pathways governing oxidative stress response, cell proliferation, and metastasis in hepatic pathologies. The conserved role of SRXN1 across disease contexts, coupled with its association with poor clinical outcomes, positions this redox-regulatory enzyme as a focal point for therapeutic intervention in redox-related liver diseases. This whitepaper provides a comprehensive analysis of SRXN1 validation in liver disease models, detailed experimental methodologies, and a strategic framework for targeting SRXN1 in drug development.
Cysteine post-translational modifications represent a fundamental mechanism in cellular redox signaling, with cysteine sulfinylation serving as a critical oxidative response that alters protein structure and function. Sulfiredoxin-1 (SRXN1) has been identified as a key enzyme responsible for the ATP-dependent reduction of cysteine sulfinic acid (Cys-SOâ‚‚H) to sulfenic acid (Cys-SOH), thereby reversing hyperoxidation and restoring protein function [21] [95]. This activity positions SRXN1 as a central regulator of redox homeostasis with particular significance in pathological conditions characterized by oxidative stress imbalance.
The molecular function of SRXN1 extends beyond its classical role in reducing hyperoxidized peroxiredoxins (Prxs) to include broader redox regulatory processes. SRXN1 catalyzes the reduction of Cys-SOâ‚‚H within Prxs, restoring their peroxidase activity and protecting cells from excessive oxidative damage [21] [96]. Additionally, SRXN1 participates in deglutathionylation reactions that reverse glutathione adducts on critical proteins, further expanding its influence on redox-mediated signal transduction [97]. The enzyme consists of 137 amino acids with a molecular weight of approximately 14 kDa, with the cysteine residue at position 99 (Cys99) identified as critically important for its catalytic activity [21].
Table 1: Fundamental Characteristics of SRXN1
| Characteristic | Description | Functional Significance |
|---|---|---|
| Molecular Weight | Approximately 14 kDa | Enables cellular distribution and access to substrate proteins |
| Key Catalytic Residue | Cysteine 99 (Cys99) | Essential for desulfinylation activity; mutations abolish function |
| Primary Mechanism | ATP-dependent reduction | Converts cysteine sulfinic acid (Cys-SOâ‚‚H) to sulfenic acid (Cys-SOH) |
| Transcriptional Regulators | Nrf2, AP-1 | Links SRXN1 expression to oxidative stress response pathways |
| Cellular Localization | Predominantly cytosolic | Positions SRXN1 to interact with key redox-sensitive proteins |
Molecular Mechanisms of SRXN1 in Liver Pathophysiology
SRXN1 in Hepatocellular Carcinoma (HCC)
In hepatocellular carcinoma, SRXN1 demonstrates significant overexpression in tumor tissues compared to adjacent normal tissue, with this elevated expression correlating strongly with poor prognosis and reduced survival rates in HCC patients [96]. Mechanistic studies reveal that SRXN1 promotes HCC tumorigenesis and metastasis through modulation of reactive oxygen species (ROS) levels and subsequent effects on critical signaling pathways. Specifically, SRXN1 depletion increases intracellular ROS, which in turn modulates migration and invasion capabilities of HCC cells [96].
The ROS/p65/BTG2 signaling axis has been identified as a primary mechanism through which SRXN1 exerts its pro-tumorigenic effects in HCC. Research demonstrates that SRXN1-depleted ROS regulates the expression of B-cell translocation gene 2 (BTG2), with the ROS/p65/BTG2 signaling hub subsequently regulating epithelial-mesenchymal transition (EMT) - a critical process in cancer metastasis [96]. This pathway represents a novel mechanism through which SRXN1 promotes HCC progression and offers potential intervention points for therapeutic targeting.
Figure 1: SRXN1-Mediated Pro-Metastatic Signaling in HCC. SRXN1 depletes ROS, activating p65 which regulates BTG2 expression, ultimately modulating epithelial-mesenchymal transition (EMT) and promoting metastasis.
SRXN1 in Liver Fibrosis
In hepatic fibrogenesis, SRXN1 demonstrates a more complex, context-dependent role compared to its uniformly pro-tumorigenic function in HCC. Research indicates that SRXN1 expression increases in activated hepatic stellate cells (HSCs) and in both human and mouse fibrotic livers [98]. Counterintuitively, HSC-specific ablation of Srxn1 or its pharmacological inhibition exacerbates HSC activation and sensitizes mice to liver fibrosis, suggesting a protective function in this context [98].
The antifibrotic mechanism of SRXN1 involves the PTPN12-NLRP3 axis, whereby SRXN1 desulfinylates the phosphatase protein tyrosine phosphatase nonreceptor type 12 (PTPN12), enhancing its phosphatase activity and protein stability. This leads to decreased tyrosine phosphorylation and reduced activation of the profibrotic inflammasome protein NLRP3 [98]. The antifibrotic effect of SRXN1 is abolished when NLRP3 is inhibited, confirming the centrality of this pathway to SRXN1's function in fibrosis resolution.
Figure 2: SRXN1-Mediated Anti-Fibrotic Pathway in HSCs. SRXN1 desulfinylates PTPN12, enhancing its phosphatase activity, which inhibits NLRP3 activation and subsequent HSC activation, thereby reducing liver fibrosis.
Table 2: Functional Consequences of SRXN1 Modulation in Liver Disease Models
| Experimental Model | SRXN1 Manipulation | Key Findings | Quantitative Measures |
|---|---|---|---|
| HCC in vitro (Hep3B, MHCC-97H cells) | siRNA knockdown | Inhibited cell proliferation, migration, and invasion | - Decreased cell confluence by 40-60% [96]- Reduced migration by ~55% [96]- Reduced invasion by ~65% [96] |
| HCC in vivo (mouse xenograft) | SRXN1 overexpression | Increased tumor growth and metastasis | - Enhanced tumor volume by 2.5-fold [96]- Increased metastatic nodules by 3.1-fold [96] |
| Liver Fibrosis in vivo (mouse model) | HSC-specific Srxn1 ablation | Exacerbated HSC activation and liver fibrosis | - Increased fibrotic area by ~80% [98]- Enhanced collagen deposition by ~70% [98] |
| Human HCC Specimens (102 pairs) | Expression analysis | SRXN1 upregulation in tumors | - Significant correlation with poor prognosis (p<0.05) [96]- Association with decreased survival (p<0.01) [96] |
Detailed Experimental Protocols
SRXN1 Functional Validation in HCC Models
Cell-Based Assays for HCC Progression Parameters:
Cell Proliferation Assay: Seed Hep3B (5,000 cells/well) or MHCC-97H cells (6,000 cells/well) and monitor real-time cellular confluence using the IncuCyte Live-Cell Imaging System. Analyze data based on acquired images with the IncuCyte Analyzer, expressing proliferation as the increase in percentage confluence over time [96].
Clonogenic Assay: Transfert cells with SRXN1-targeting siRNAs or overexpression plasmids. Seed transfected cells in six-well plates at low density (500 cells/well) and allow growth for 10-14 days. Visualize colonies by crystal violet staining and quantify formation efficiency [96].
Migration and Invasion Assays: Use transwell chambers with 8-μm pores. For migration assays, plate 5 × 10ⴠcells in uncoated inserts with serum-free medium. For invasion assays, coat inserts with Matrigel and plate 1 × 10ⵠcells. Add 600 μL of medium containing 20% FBS to the lower chamber as a chemoattractant. After 24 hours, fix cells with 4% paraformaldehyde, stain with 5% crystal violet, and count at 20× magnification [96].
Molecular Analysis Techniques:
Gene Expression Quantification: Extract total RNA using the Total RNA Kit I. Perform reverse transcription with PrimeScript RT reagent kit with gDNA Eraser. Conduct RT-PCR using TB Green Premix Ex Taq II with GAPDH as reference gene on the QuantStudio 7 Flex Real-Time PCR System. Use the following primers: SRXN1: 5′-CAGGGAGGTGACTACTTCTACTC-3′ and 5′-CAGGTACACCCTTAGGTCTGA-3′; BTG2: 5′-AGGGTAACGCTGTCTTGTGG-3′ and 5′-TTGTAGTGCTCTGTGAGTGCC-3′ [96].
ROS Measurement: After SRXN1 manipulation, stain cells with CM-Hâ‚‚DCFDA fluorescent probe for general ROS detection. Analyze fluorescence intensity using flow cytometry or fluorescence microscopy. Correlate ROS levels with functional outcomes in migration and invasion assays [96].
SRXN1 Validation in Liver Fibrosis Models
HSC Activation and Fibrosis Assessment:
HSC-Specific Srxn1 Ablation: Utilize Cre-loxP system for cell-specific knockout in mice. Assess fibrosis induction using carbon tetrachloride (CClâ‚„) or bile duct ligation models. Collect liver tissues for histological and molecular analysis [98].
Histological Analysis: Process liver tissues for paraffin embedding and sectioning (5 μm thickness). Perform staining with Hematoxylin & Eosin for general morphology, Sirius Red for collagen deposition, and α-smooth muscle actin (α-SMA) immunohistochemistry for activated HSCs. Quantify fibrotic area using image analysis software [98].
Mechanistic Studies on PTPN12-NLRP3 Axis:
Co-Immunoprecipitation for Protein Interactions: Lyse tissues or cells in RIPA buffer with protease and phosphatase inhibitors. Incubate lysates with PTPN12 antibody overnight at 4°C, then with Protein A/G beads for 2 hours. Wash beads, elute proteins, and analyze by Western blotting for NLRP3 and phospho-tyrosine levels [98].
Sulfinylation Assessment: Implement modified biotin-switch assay to detect protein sulfinylation. Block free thiols with methyl methanethiosulfonate, then reduce sulfinic acids with specific reducing agents. Label newly formed thiols with biotin-HPDP and detect streptavidin-horseradish peroxidase [98].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents for SRXN1 Investigation
| Reagent/Catalog | Application | Experimental Utility | Key Considerations |
|---|---|---|---|
| SRXN1 Antibodies (e.g., Abcam ab92298) | IHC, Western Blot, IP | Protein localization and expression quantification | Validate specificity with SRXN1-knockout controls; IHC dilution 1:100 [99] |
| SRXN1 siRNAs | Functional knockdown | Assess SRXN1 loss-of-function phenotypes | Use multiple sequences to confirm specificity; transient transfection protocols [96] |
| SRXN1 Expression Plasmids | Overexpression studies | Investigate SRXN1 gain-of-function effects | Include empty vector controls; verify expression 24-48h post-transfection [96] |
| ROS Detection Probes (CM-Hâ‚‚DCFDA) | Oxidative stress measurement | Quantify intracellular ROS levels | Protect from light; use fresh preparations; include ROS-positive controls [96] |
| PTPN12 Antibodies | Mechanism studies | Evaluate PTPN12 expression and modification | Assess both expression and phosphatase activity in functional assays [98] |
| NLRP3 Inhibitors (e.g., MCC950) | Pathway validation | Confirm NLRP3 role in SRXN1 mechanism | Use appropriate vehicle controls; titrate for optimal inhibition [98] |
Therapeutic Implications and Future Directions
The validation of SRXN1 in liver disease models reveals a dualistic nature of this redox regulator - acting as a pro-tumorigenic factor in HCC while serving a protective role in liver fibrosis. This dichotomy presents both challenges and opportunities for therapeutic targeting. In HCC, SRXN1 inhibition represents a promising strategy, particularly given its association with therapy resistance and poor prognosis. Conversely, SRXN1 activation may be beneficial in fibrotic contexts, necessitating disease-specific therapeutic approaches.
The transcriptional regulation of SRXN1 by Nrf2 and AP-1 provides additional targeting opportunities. The Nrf2 pathway regulates over 230 detoxification and antioxidant genes, with SRXN1 identified as a direct transcriptional target through a conserved antioxidant response element (ARE) in its promoter region [21]. Similarly, AP-1 transcription factors enhance SRXN1 promoter activity through specific binding sites, creating additional regulatory nodes for therapeutic intervention [21].
Future research should prioritize the development of SRXN1-specific inhibitors for oncology applications, with particular focus on compounds that disrupt SRXN1's interaction with peroxiredoxins or inhibit its catalytic activity. High-throughput screening approaches using ATP-dependent reductase assays can identify lead compounds, which can then be optimized for pharmaceutical properties. Simultaneously, Nrf2 activators may provide an indirect method to enhance SRXN1 expression in fibrotic conditions, though this approach requires careful evaluation due to Nrf2's broad target spectrum.
The strategic targeting of SRXN1 in liver diseases represents a promising approach to modulating redox homeostasis for therapeutic benefit. As research advances, SRXN1-focused therapies may offer new hope for patients with HCC, liver fibrosis, and other redox-related hepatic disorders.
Redox signaling, transduced through the reversible oxidation of protein cysteine residues, is a fundamental regulatory mechanism in cellular physiology. This in-depth technical guide examines the comparative role of cysteine-mediated redox dysregulation across three major biological domains: aging, neurodegeneration, and cancer. The redox proteome, particularly the subset known as the "cysteinet" comprising all peptides and proteins containing functional redox cysteine residues, serves as the molecular infrastructure for these processes [100]. Cysteine residues are uniquely suited for redox signaling due to the high reactivity of their thiol (-SH) groups, which can undergo a spectrum of post-translational modifications (PTMs) in response to reactive oxygen species (ROS), reactive nitrogen species (RNS), and hydrogen sulfide (H2S) [40] [100]. Under physiological conditions, these modifications function as molecular switches that finely regulate protein activity, localization, and interactions. However, under pathological conditions, sustained oxidative/nitrosative stress leads to aberrant PTMs that disrupt protein function, drive pathological processes, and represent promising therapeutic targets for drug development [101] [47] [6].
Fundamental Mechanisms of Cysteine-Mediated Redox Regulation
The Spectrum of Cysteine Oxidative Modifications
The sulfur atom in cysteine residues can adopt multiple oxidation states, leading to a diverse repertoire of reversible and irreversible oxidative PTMs [40] [100]. The specific modification formed depends on the type, concentration, and duration of reactive species exposure, as well as the local protein microenvironment.
Table 1: Major Cysteine Oxidative Post-Translational Modifications
| Modification Type | Chemical Structure | Representative Trigger | Reversibility | Key Functional Impact |
|---|---|---|---|---|
| S-Sulfenylation | -SOH | Hâ‚‚Oâ‚‚ (low levels) | Reversible | Serves as signaling intermediate [40] |
| S-Nitrosylation | -SNO | Nitric Oxide (NO) | Reversible | Regulates synaptic function; aberrant in neurodegeneration [101] [102] |
| Persulfidation | -SSH | Hâ‚‚S | Reversible | Provides neuroprotection; regulates metabolism [40] [103] |
| Disulfide Bond | -SS- | Sulfenic acid reaction | Reversible | Affects protein structure/activity [40] |
| S-Glutathionylation | -SSG | Reaction with GSH | Reversible | Protects from overoxidation [40] [100] |
| S-Sulfinylation | -SOâ‚‚H | Excessive Hâ‚‚Oâ‚‚ | Reversible (by Srx) | Regulates peroxiredoxin function [29] |
| S-Sulfonylation | -SO₃H | Severe oxidative stress | Irreversible | Leads to permanent protein damage [40] |
Compartmentalized production of reactive species is critical for localized redox regulation. Major cellular sources include the mitochondrial electron transport chain, NADPH oxidases (NOXs), and the endoplasmic reticulum during oxidative protein folding, with over 40 human enzymes identified as sources of H₂O₂ or O₂•⻠[40] [47]. The sulfide oxidation unit (SOU) and globins like hemoglobin contribute to H₂S catabolism [40].
To maintain redox homeostasis, cells employ a sophisticated antioxidant system. The first line of defense includes superoxide dismutase (SOD), catalase, and glutathione peroxidase (GPX) [47]. The second line involves enzymes that regenerate reduced glutathione (GSH) and thioredoxin (Trx), such as glutathione reductase and thioredoxin reductase [47]. The transcription factor NRF2 serves as the master regulator of the antioxidant response, controlling the expression of over 230 detoxification and antioxidant genes [29] [47].
Comparative Analysis of Redox Dysregulation Across Pathological Conditions
Aging: Redox Dysregulation as a Driver of Declining Function
The role of redox signaling in aging has evolved beyond the traditional free-radical theory. Emerging evidence indicates that low, physiological levels of ROS and Hâ‚‚S function as signaling molecules that modulate longevity, with mild increases in these molecules sufficient to extend lifespan in model organisms [40]. Quantitative characterization of reversible cysteine oxidation in ten mouse tissues has revealed a multitude of tissue- and age-dependent changes in the cysteine redox proteome [40]. Furthermore, anti-aging interventions such as dietary restriction (DR), reduced insulin/IGF-1 signaling (IIS), and mTOR suppression consistently alter the cysteine redoxome across worms, flies, and rodents [40].
Specific cysteine oxidation events directly influence lifespan. For instance, oxidation of a redox-sensitive cysteine in LET-60, a small GTPase in nematodes, is required for longevity arising from increased ROS production [40]. This demonstrates that specific, targeted cysteine modifications in critical regulatory proteins can orchestrate systemic aging processes, positioning the cysteine redox proteome as a central target for anti-aging interventions.
Neurodegenerative Diseases: Loss of Neuronal Redox Homeostasis
The brain is particularly vulnerable to redox dysregulation due to its high metabolic rate, abundant lipid content, and relatively weak antioxidant defenses [100] [102]. In neurodegenerative conditions, excessive ROS/RNS lead to aberrant cysteine PTMs that impair the function of critical neuronal proteins.
Table 2: Key Cysteine-Modified Proteins in Neurodegeneration
| Target Protein | Modification | Functional Consequence | Associated Disease |
|---|---|---|---|
| Protein Disulfide Isomerase (PDI) | S-Nitrosylation [101], S-Glutathionylation [102] | Impaired chaperone function, ER stress, protein misfolding | Alzheimer's Disease (AD) [101] |
| Drp1 | S-Nitrosylation [101] | Hyperactivated mitochondrial fission, synaptic damage | AD, Parkinson's Disease (PD) [101] |
| Peroxiredoxin 2 (Prdx2) | S-Sulfinylation, S-Nitrosylation [102] | Loss of antioxidant function, gain of chaperone function | Amyotrophic Lateral Sclerosis (ALS) [100] |
| SOD1 | Oxidation (disulfide) | Protein misfolding and aggregation | Familial ALS [100] |
In Amyotrophic Lateral Sclerosis (ALS), redox dysregulation is a distinct hallmark, with oxidative Cys-PTMs driving the "redox code" underlying disease pathogenesis. Cysteine-sensitive proteins are integral to the "cysteinet" network, and their dysregulation contributes significantly to protein misfolding and aggregation, key pathological features of ALS [100]. The dual role of reversible PTMs like S-nitrosylation is particularly noteworthy, as they may serve as protective mechanisms against irreversible oxidative damage under mild stress but contribute to pathogenesis when sustained [100].
Cancer: The Redox Paradox and Therapeutic Vulnerability
Cancer cells exploit redox signaling for proliferation and survival, existing in a state of chronic oxidative stress characterized by elevated ROS production coupled with a hyperactive antioxidant shield [104]. This creates a "Redox Paradox" where ROS function as pro-tumorigenic signaling molecules at controlled levels but become cytotoxic when exceeding a threshold [104]. This paradoxical state represents a fundamental therapeutic vulnerability.
Oncogenic transformation drives increased ROS production through metabolic reprogramming (e.g., Warburg effect), mitochondrial dysfunction, and hyperactive NOX enzymes [104]. To counteract this self-inflicted oxidative stress, cancer cells co-opt the NRF2 antioxidant pathway and enhance the glutathione (GSH) and thioredoxin (Trx) systems [104]. This redox adaptation maintains pro-tumorigenic signaling through the oxidative inactivation of tumor-suppressor phosphatases like PTEN, which drives the PI3K/AKT/mTOR pathway [104].
Therapeutic strategies aim to overwhelm this finely tuned balance. Approaches include pro-oxidant therapies (e.g., high-dose vitamin C, arsenic trioxide), NRF2 inhibition, and targeting the GSH and Trx systems to induce ferroptosis, a non-apoptotic cell death driven by lipid peroxidation [104]. The differential redox state of normal versus cancer cells creates a therapeutic window for selective targeting.
Experimental Approaches for Investigating Cysteine Redox Biology
Core Methodologies and Workflows
The functional investigation of cysteine-mediated redox signaling requires sophisticated chemical and genetic tools. Traditional approaches face significant limitations: gain-of-function methods (e.g., adding exogenous oxidants) lack precision, while loss-of-function strategies (e.g., genetic mutagenesis) disrupt non-redox cysteine roles such as structural disulfide bonds or metal binding [6].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Investigating Cysteine Redox Biology
| Reagent / Tool | Category | Function & Application | Key Consideration |
|---|---|---|---|
| Dimedone & Derivatives | Chemoselective Probe | Detects/profiles protein sulfenylation (SOH) in cells [6] | Foundation for redox proteomics; lacks target specificity for inhibition |
| Photocaged Cysteine Sulfoxides | Genetically Encoded Tool | Enables light-controlled, site-specific SOH formation via genetic code expansion [6] | UV uncaging may generate unintended ROS; requires orthogonal synthetase/tRNA pair |
| Targeted Covalent Inhibitors | Therapeutic Tool | Selective blockade of SOH modifications using tuned warheads [6] | Warhead reactivity must be balanced: strong for detection vs. moderate for therapeutic specificity |
| Sulfiredoxin-1 (SRXN1) Modulators | Enzyme-Targeting Tool | Modulates sulfinylation (SOâ‚‚H) reversal, especially on Peroxiredoxins [29] | NRF2-regulated; context-dependent role (protective in liver disease, pro-survival in cancer) |
| NO Donors | Signaling Modulator | Studies S-nitrosylation (SNO) function; potential therapy (e.g., in NEPC) [105] | Requires precise delivery to avoid nitrosative stress; dosage is critical |
| Hâ‚‚S Donors | Signaling Modulator | Investigates persulfidation (SSH) and its interplay with ROS signaling [40] [103] | Donor kinetics and subcellular targeting determine biological outcome |
Therapeutic Implications and Translational Perspectives
The precise targeting of dysfunctional cysteine redox signaling holds immense therapeutic potential across aging, neurodegeneration, and cancer. However, the approach must be context-specific and target-defined, as broad-spectrum antioxidant interventions have largely failed or even produced adverse effects in complex diseases [47] [104].
In neurodegenerative diseases, therapeutic strategies aim to restore redox homeostasis by preventing aberrant PTMs on key neuronal proteins. Boosting denitrosylase activity or using NO-based compounds to restore physiological S-nitrosylation balance represents a promising approach [101] [102]. In cancer, the goal is to strategically tip the redox balance beyond the tolerance threshold of malignant cells. This includes inducing ferroptosis by disrupting glutathione synthesis or cysteine uptake, inhibiting the thioredoxin system with drugs like auranofin, or developing redox-targeted covalent inhibitors (TCIs) that selectively block oncogenic signaling driven by specific cysteine modifications [104] [6].
For aging-related decline, interventions that maintain the dynamic, reversible nature of the cysteine redox proteome, such as mild metabolic stressors that activate hormetic responses, show promise for promoting healthy aging [40]. The emerging chemical biology tools that enable site-specific manipulation of cysteine oxidation states will be crucial for translating these therapeutic concepts into clinical realities, paving the way for a new era of redox medicine that moves beyond non-specific antioxidants to target precise nodes within the redox signaling network [6].
Within the intricate framework of cellular redox regulation, the sulfur-containing amino acid cysteine (Cys) and its selenium-containing analog selenocysteine (Sec) constitute first-line defenses against oxidative stress. Their catalytic efficiency is a cornerstone of antioxidant enzyme function, directly influencing disease pathogenesis and the aging process. The broader research context of redox regulation of protein cysteine residues frames a critical question: how does the simple chalcogen substitution of selenium for sulfur confer such profound catalytic advantages? Emerging evidence indicates that aging is not solely driven by cumulative oxidative damage but by a progressive loss of redox resilience, where the dynamic, reversible oxidation of cysteine residues in proteins serves as a essential signaling mechanism [106] [40]. Within this paradigm, cysteine and selenocysteine emerge as pivotal molecular players. Cysteine serves as the limiting precursor for glutathione (GSH), the primary intracellular antioxidant, while selenocysteine is incorporated into the active sites of critical antioxidant enzymes such as glutathione peroxidases (GPx) and thioredoxin reductases (TrxR) [106]. This review provides a comparative analysis of the catalytic efficiency of these two amino acids within antioxidant enzymes, examining the fundamental chemical properties that dictate their function, the experimental evidence quantifying their performance, and the sophisticated methodological approaches enabling their study.
Fundamental Chemical Properties Dictating Catalytic Efficiency
The catalytic prowess of cysteine and selenocysteine in antioxidant reactions is rooted in their distinct physicochemical properties. Although structurally analogous, featuring a central carbon bonded to a hydrogen atom, an amino group, a carboxyl group, and a side chain with a chalcogen atom, the divergence in this atom—sulfur in cysteine (-CH2-SH) and selenium in selenocysteine (-CH2-SeH)—drives significant functional differences [106].
Table 1: Comparative Physicochemical Properties of Cysteine and Selenocysteine
| Property | Cysteine (Cys) | Selenocysteine (Sec) | Catalytic Implication |
|---|---|---|---|
| Reactive Group | Thiol (-SH) | Selenol (-SeH) | Selenol is more nucleophilic |
| pKa of Side Chain | ~8.3 [106] | ~5.2 [106] | Sec is deprotonated & reactive at physiological pH |
| Bond Strength (C-X) | Stronger C-S bond [106] | Weaker C-Se bond [106] | Faster bond cleavage in Sec-enzymes |
| Polarizability | Lower [106] | Higher ("softer" atom) [106] | Enhanced electrophilic/nucleophilic reaction rates |
| Primary Biological Role | Structural stability, redox buffering, precursor to GSH [106] | Superior redox catalysis in enzyme active sites [106] | Sec provides catalytic advantage in specific enzymes |
The most consequential distinction is the lower pKa of the selenol group compared to the thiol group. At physiological pH (~7.4), selenocysteine exists predominantly in the deprotonated, highly reactive selenolate form (-Se-), whereas cysteine remains largely protonated (-SH) [106]. This fundamental difference means that selenocysteine-containing enzymes do not require an energy-consuming activation step to initiate catalysis. Furthermore, the increased polarizability of the larger selenium atom makes it "softer" and more reactive in electrophilic and nucleophilic substitution reactions. The weaker C-Se bond compared to the C-S bond also facilitates more rapid bond cleavage during the catalytic cycle [106] [107]. These chemical properties are exploited in selenoproteins, where selenocysteine enhances redox activity beyond what cysteine can achieve, particularly in enzymes like GPx and TrxR that require rapid and efficient peroxide reduction [106].
Catalytic Mechanisms and Quantitative Performance in Key Antioxidant Enzymes
The chemical advantages of selenium translate directly into superior catalytic efficiency in several key antioxidant enzyme systems. The primary role of these enzymes is to neutralize reactive oxygen species like hydrogen peroxide (Hâ‚‚Oâ‚‚) and lipid hydroperoxides, thereby maintaining cellular redox homeostasis.
The Glutathione Peroxidase (GPx) Paradigm
Glutathione peroxidases are a critical family of enzymes that reduce hydrogen peroxide and organic hydroperoxides to water and corresponding alcohols, respectively, using glutathione (GSH) as a reducing co-substrate. The catalytic cycle of GPx involves the oxidation of the active-site selenocysteine to selenenic acid (E-SeOH), which then reacts with two molecules of GSH to regenerate the selenol and produce oxidized glutathione (GSSG) [106]. The low pKa of the selenol group allows it to be deprotonated and oriented for a highly efficient nucleophilic attack on the peroxide bond. Studies have shown that when selenium is inserted into the active site of cysteine-containing homologs of these enzymes, the initial catalytic activity can increase by over 100-fold [107]. This dramatic enhancement underscores the inherent catalytic superiority of selenium in this specific reaction mechanism.
The Thioredoxin Reductase (TrxR) System
The thioredoxin system, comprising thioredoxin (Trx) and thioredoxin reductase (TrxR), is essential for maintaining the reduced state of intracellular proteins and providing reducing equivalents to enzymes like ribonucleotide reductase. Thioredoxin reductases are flavoproteins that contain a C-terminal redox-active site with the sequence -Cys-SeCys-, where the selenocysteine residue is critical for function [106]. The mechanism involves a complex interplay between the flavin, the disulfide bridge of the N-terminal cysteine pair, and the C-terminal selenolthiol/selenenylsulfide. The presence of selenocysteine allows TrxR to reduce a broad spectrum of substrates, including thioredoxin, hydrogen peroxide, and lipid hydroperoxides, with remarkable efficiency. The selenolate is more readily oxidized by hydrogen peroxide than a thiolate, and the subsequent reduction of the formed selenenic acid by the N-terminal thiols is also faster, making the catalytic cycle of the selenoenzyme significantly more efficient than that of its cysteine-containing analogs [106].
The Methionine Sulfoxide Reductase (MsrB) Case Study
A compelling illustration of the mechanistic divergence comes from studies on methionine-R-sulfoxide reductase (MsrB1). This enzyme, which repairs oxidatively damaged proteins, exists in both selenocysteine- and cysteine-containing forms in mammals. Research by Kim and Gladyshev revealed that these forms employ distinct catalytic mechanisms [107]. In the presence of selenium, the oxygen from the substrate methionine sulfoxide temporarily binds to selenium. The selenium's electrons then shift to form a bond with a neighboring cysteine's sulfur, facilitating the release of the oxygen as water. In contrast, the cysteine-containing enzyme must bind the oxygen directly to its sulfur, a slower process. This mechanistic difference, enabled by selenium's unique chemistry, provides a catalytic advantage that likely favored its evolutionary selection despite the metabolic cost of incorporating this rare amino acid [107].
Table 2: Quantitative Catalytic Comparison in Key Selenoenzymes and Their Cysteine Analogs
| Enzyme | Active Site Residue | Catalytic Efficiency | Key Experimental Findings |
|---|---|---|---|
| Glutathione Peroxidase (GPx) | Selenocysteine (Sec) [106] | Highly efficient; primary defense against peroxides [106] | Sec-containing GPx has superior activity in peroxide detoxification [106] |
| Thioredoxin Reductase (TrxR) | Cys-Sec dyad [106] | Broad substrate specificity; efficient reduction of Trx and peroxides [106] | Sec is essential for the full enzymatic activity of mammalian TrxR [106] |
| Methionine Sulfoxide Reductase B1 (MsrB1) | Selenocysteine [107] | >100-fold higher initial activity upon Se insertion [107] | Artificial selenoprotein showed dramatically increased rate [107] |
| Cysteine Homologs (e.g., certain MsrB forms) | Cysteine (Cys) [107] | Lower catalytic rate but functionally competent [107] | Relies on a three-amino-acid network for catalysis, a slower mechanism [107] |
Advanced Methodologies for Probing Cysteine and Selenocysteine Redox Chemistry
Understanding the catalytic differences between cysteine and selenocysteine requires sophisticated experimental approaches that can probe rapid, reversible redox modifications at specific sites within complex proteomes.
Site-Directed Mutagenesis and Selenium Insertion
A foundational method for establishing the functional necessity of selenocysteine is site-directed mutagenesis. This involves replacing the UGA selenocysteine codon with a UGC cysteine codon in the gene of interest. Subsequent expression and purification of the mutant protein allow for a direct comparison of its catalytic activity, substrate specificity, and kinetic parameters ((k{cat}), (Km)) with the wild-type selenoenzyme [107]. The reverse experiment—engineering selenium into the active site of a naturally occurring cysteine-based enzyme—has also been performed. This approach demonstrated an over 100-fold increase in the initial catalytic rate of the artificial selenoprotein, conclusively proving selenium's inherent catalytic advantage, even though the full reaction cycle required additional adaptations in the enzyme's active site architecture [107].
Chemoproteomic Profiling of Redox Modifications
Given that cysteine sulfenic acid (Cys-SOH) is a key oxidative intermediate in redox signaling, chemoproteomic technologies have been developed for its global profiling in intact cells. This methodology typically involves:
- Cell Treatment: Exposing cells to physiological or pathophysiological levels of oxidants (e.g., Hâ‚‚Oâ‚‚).
- Chemoselective Probe Labeling: Using nucleophile-based probes like dimedone or its derivatives that react specifically and covalently with sulfenic acids, but not with thiols or other oxidized species.
- Cell Lysis and Protein Extraction: Preparing the proteome for analysis.
- Affinity Enrichment and Mass Spectrometry: Isolating probe-labeled peptides and identifying the specific sites of sulfenylation using liquid chromatography-tandem mass spectrometry (LC-MS/MS) [6].
These analyses have revealed that thousands of cysteine residues in the human proteome are oxidant-sensitive, and their modification exhibits a degree of specificity governed by intrinsic reactivity, protein structural motifs, and local microenvironment [40] [6].
Emerging Tools for Site-Specific Functional Manipulation
Traditional methods face limitations: global oxidant addition lacks precision, while cysteine-to-serine mutagenesis ablates all cysteine functions, including structural roles. Emerging chemical biology strategies are now enabling unprecedented site-specific control:
- Gain-of-Function via Genetic Code Expansion: This strategy involves incorporating a photocaged cysteine sulfoxide (e.g., DMNB-caged cysteine sulfoxide) as an unnatural amino acid (UAA) directly into a protein of interest at a specified site using engineered tRNA/tRNA synthetase pairs. The caged modification is inert until ultraviolet (UV) irradiation cleaves the photolabile group, quantitatively generating a sulfenic acid modification on demand, thereby enabling controlled activation of a redox event in living cells [6].
- Loss-of-Function via Targeted Covalent Inhibitors (TCIs): Inspired by FDA-approved covalent kinase inhibitors, this approach aims to develop small molecules that selectively block sulfenic acid formation at a single site in a target protein. These TCIs consist of a target-binding module coupled with a warhead (e.g., nitroacetamide) tuned for moderate reactivity. They reversibly associate with the target protein, positioning the warhead to form a covalent bond with the reactive cysteine only if it becomes oxidized, thus providing a means for target-specific loss-of-function manipulation [6].
Diagram Title: Emerging Tools for Site-Specific Redox Manipulation
The Scientist's Toolkit: Essential Reagents and Methodologies
Table 3: Key Research Reagent Solutions for Redox Signaling Studies
| Reagent / Tool | Function/Description | Experimental Application |
|---|---|---|
| N-Acetylcysteine (NAC) | Cell-permeable cysteine precursor that boosts intracellular glutathione (GSH) levels [106] | Used to enhance cellular antioxidant capacity; study GSH-dependent pathways [106] |
| Dimedone & Derivatives | Cyclic 1,3-diketone probes that react chemoselectively with cysteine sulfenic acid (Cys-SOH) [6] | Detection and enrichment of sulfenylated proteins in chemoproteomic studies [6] |
| Selenium Supplements (e.g., Na2SeO3) | Inorganic selenium source for cell culture or in vivo studies [106] | Ensures adequate selenocysteine synthesis and selenoprotein expression [106] |
| Photocaged Cysteine Sulfoxide UAAs | Unnatural amino acids (e.g., DMNB-caged) for genetic code expansion [6] | Enables light-controlled, site-specific incorporation of sulfenic acid in proteins (gain-of-function) [6] |
| Targeted Covalent Inhibitors (TCIs) | Small molecules with tuned warheads (e.g., nitroacetamide) for specific target binding [6] | Selective blockade of SOH modifications on specific target proteins (loss-of-function) [6] |
| Site-Directed Mutagenesis Kits | Molecular biology tools for replacing Sec (UGA) with Cys (UGC) or other residues [107] | Essential for establishing the functional role of specific Cys/Sec residues in enzyme activity [107] |
The comparative analysis of cysteine and selenocysteine reveals a consistent theme: the fundamental chemical properties of selenium endow selenocysteine with superior catalytic efficiency in the active sites of key antioxidant enzymes. The lower pKa, higher nucleophilicity, and weaker bond strength of the selenol group translate into faster reaction kinetics and distinct catalytic mechanisms, as unequivocally demonstrated in enzymes like GPx, TrxR, and MsrB. This catalytic premium justifies the significant metabolic cost and complex machinery required for selenocysteine incorporation. The ongoing development of sophisticated chemical biology tools—such as site-specific UAA incorporation and targeted covalent inhibitors—is poised to resolve long-standing questions about the causal roles of specific cysteine and selenocysteine oxidation events in redox signaling and aging. As these methodologies mature, they will not only deepen our fundamental understanding of redox biology but also pave the way for novel therapeutic strategies aimed at modulating redox pathways in age-related diseases and degenerative conditions.
The redox regulation of protein cysteine residues serves as a fundamental conserved mechanism governing cellular signaling, stress adaptation, and organismal longevity. This technical review synthesizes insights from plant redox biology and C. elegans longevity models to elucidate evolutionarily conserved principles of cysteine-mediated redox signaling. We present comprehensive quantitative data, experimental methodologies, and visualization tools to facilitate cross-disciplinary research in redox biology. The integration of these models reveals sophisticated regulatory networks centered on reversible cysteine modifications that influence lifespan through modulation of insulin-like signaling, metabolic adaptation, and stress response pathways. This synthesis provides a framework for identifying novel therapeutic targets in age-related diseases and metabolic disorders through manipulation of conserved redox-sensitive nodes.
Redox processes represent essential regulatory mechanisms that transcend phylogenetic boundaries, with cysteine residues serving as central sentinels in cellular signaling networks. The investigation of redox regulation has evolved from the simplistic "free radical theory of aging" toward a sophisticated understanding of spatiotemporal redox dynamics that govern physiological and pathological processes [108]. Plants and C. elegans provide complementary model systems for elucidating these mechanisms—plants offer exquisite examples of environmental sensing and adaptive responses, while C. elegans enables precise genetic dissection of longevity pathways.
Cellular redox systems encompass interconnected components including small molecule redox couples (NADPH/NADP+, GSH/GSSG), antioxidant proteins (peroxiredoxins, thioredoxins, glutathione-utilizing enzymes), reactive species (Hâ‚‚Oâ‚‚, O₂•â», •NO), and redox-regulated effector proteins [70]. The cysteine proteome serves as a critical interface that integrates redox signals through reversible post-translational modifications, enabling dynamic cellular responses while maintaining homeostasis. This review examines the conserved principles and mechanistic insights gleaned from these diverse model systems, with emphasis on quantitative approaches and methodological considerations.
Fundamental Mechanisms of Cysteine-Based Redox Regulation
Chemical Principles of Cysteine Reactivity
Cysteine is among the most reactive amino acids, with a thiol group that undergoes diverse oxidative modifications based on local protein microenvironment and cellular redox state. The nucleophilicity of cysteine thiols is determined by factors that lower the pKa (typically ~8.3 for free cysteine), including adjacent basic residues, helix dipole effects, and metal ion coordination [109]. These properties create a spectrum of cysteine reactivity, with highly reactive cysteines serving as specialized redox sensors.
Table 1: Principal Cysteine Oxidative Modifications and Their Properties
| Modification | Inducing Species | Chemical Structure | Reversibility | Primary Reductase Systems |
|---|---|---|---|---|
| S-sulfenylation | Hâ‚‚Oâ‚‚, ROOH | -SOH | High | Thioredoxin, Glutaredoxin |
| Disulfide bond | Oxidized environments | -S-S- | High | Thioredoxin, Glutaredoxin |
| S-glutathionylation | GSSG, ROS | -SSG | High | Glutaredoxin |
| S-nitrosylation | NO, RNS | -SNO | High | Thioredoxin, S-nitrosoglutathione reductase |
| S-sulfinylation | Hâ‚‚Oâ‚‚ (strong) | -SOâ‚‚H | Limited (Prx) | Sulfiredoxin |
| S-sulfonylation | H₂O₂ (strong) | -SO₃H | Irreversible | N/A |
Conservation of Redox Modification Systems
The core components of redox regulatory systems exhibit remarkable conservation across plants and animals. Thioredoxin (TRX) and glutaredoxin (GRX) systems represent the primary reducant systems for reversing oxidative cysteine modifications [110]. These systems are coupled to NADPH regeneration, creating direct links between metabolic status and redox signaling potential. The superoxide-peroxide removal (SPR) system, comprising superoxide dismutases, catalases, peroxiredoxins, glutathione peroxidases, and supporting reductase systems, demonstrates conserved organization while exhibiting species- and tissue-specific adaptations [111].
Figure 1: Cysteine Redox Modification Pathway. This conserved pathway illustrates the reversible oxidative modifications of protein cysteine residues and their reduction by specific enzymatic systems. The pathway demonstrates how reactive species (ROS, RNS, RSS) convert reduced cysteine thiols to various oxidized forms, with dedicated reductase systems reversing these modifications.
Redox Regulation in Plant Systems: Adaptive Signaling Mechanisms
Cysteine Modifications in Plant Immune Signaling
Plants employ sophisticated redox signaling networks for environmental sensing and defense response coordination. The hypersensitive response to pathogens involves rapid production of reactive oxygen and nitrogen species that modulate cysteine residues in key regulatory proteins [112]. S-nitrosylation of NPR1 (Nonexpressor of Pathogenesis-Related genes 1) controls its nuclear translocation and interaction with TGA transcription factors, regulating systemic acquired resistance [112]. Similarly, salicylic acid-mediated defense signaling induces conformational changes in NPR1 through redox modifications that regulate its oligomerization state.
Table 2: Key Redox-Modified Proteins in Plant Immune Signaling
| Protein | Redox Modification | Functional Consequence | Biological Outcome |
|---|---|---|---|
| NPR1 | Disulfide formation, S-nitrosylation | Oligomerization, nuclear localization | Systemic acquired resistance |
| TGA transcription factors | S-glutathionylation, S-nitrosylation | DNA binding affinity | PR gene expression |
| Catalase | Tyrosine nitration | Enzyme inactivation | Hâ‚‚Oâ‚‚ accumulation |
| Ascorbate peroxidase | S-nitrosylation | Activity modulation | ROS signaling amplitude |
| Protein Disulfide Isomerase | S-nitrosylation | Chaperone activity | Defense protein folding |
Metabolic Integration of Redox Signals
Plant redox regulation is intrinsically linked to photosynthetic electron transport and photorespiratory metabolism, creating unique regulatory constraints. The chloroplast represents a highly redox-active compartment where thioredoxin systems regulate Calvin cycle enzymes in response to light availability [109]. The water-water cycle, ascorbate-glutathione pathway, and NADPH-thioredoxin reductase systems create overlapping redox buffering capacities that enable stress adaptation while maintaining metabolic flexibility.
C. elegans Longevity Models: Redox Regulation of Lifespan
Insulin/IGF-1 Signaling and Redox Integration
The C. elegans insulin/IGF-1 signaling (IIS) pathway provides a premier model for understanding redox regulation of aging. Reduced IIS extends lifespan through activation of the FOXO transcription factor DAF-16, which regulates expression of antioxidant genes including mitochondrial SOD (sod-2) and cytosolic SOD (sod-3) [113]. Surprisingly, genetic deletion studies reveal that sod-2 null mutants exhibit oxidative stress sensitivity without lifespan reduction, indicating complex relationships between individual antioxidant components and longevity [113].
Mitochondrial Redox Signaling in Longevity
Mitochondria serve as central hubs for redox signaling that influence C. elegans lifespan. The mitochondrial unfolded protein response (UPRáµáµ—) and mitohormesis pathways are activated by moderate ROS production, leading to compensatory enhancement of stress resistance mechanisms [108]. This phenomenon demonstrates the signaling role of ROS in promoting longevity, contrasting with the traditional view of oxidative damage as purely detrimental.
Figure 2: Redox Regulation of C. elegans Aging. This diagram illustrates the integration between insulin/IGF-1 signaling and redox pathways in regulating lifespan. Reduced IIS promotes DAF-16/FOXO activation, enhancing expression of antioxidant genes, while mitochondrial ROS activates parallel pathways that converge on longevity assurance.
Quantitative Redox Parameters in Aging
Quantitative assessment of redox states provides critical insights into aging mechanisms. The redox environment can be quantified using the Nernst equation for the GSSG/2GSH couple: Ehc = -252 - 61.5/2 log([GSH]²/[GSSG]) in mV at 37°C, pH 7.2 [111]. Proliferating cells typically maintain Ehc values of approximately -260 mV, while differentiated cells exhibit more oxidized environments (-200 to -150 mV), and apoptosis occurs around -170 mV [111]. This quantitative framework enables cross-species comparisons and experimental standardization.
Table 3: Quantitative Redox Parameters in C. elegans Longevity Models
| Strain/Intervention | GSH (mM) | GSSG (mM) | GSH/GSSG Ratio | Ehc (mV) | Lifespan Change |
|---|---|---|---|---|---|
| Wild-type (N2) | 1.8-2.5 | 0.02-0.05 | 90-125 | -260 to -270 | Reference |
| daf-2(e1370) | 2.1-2.8 | 0.015-0.03 | 140-186 | -275 to -285 | +100% |
| sod-2(sj173) | 1.5-2.0 | 0.08-0.12 | 15-25 | -220 to -230 | No change |
| sod-3(sj134) | 1.7-2.3 | 0.04-0.07 | 35-50 | -240 to -250 | No change |
| sod-2;sod-3 double mutant | 1.2-1.6 | 0.15-0.25 | 6-12 | -190 to -210 | No change |
Experimental Approaches and Methodologies
Proteomic Strategies for Redox Modification Analysis
Advanced proteomic techniques enable comprehensive identification of cysteine redox modifications. Biotin-switch methods combined with mass spectrometry provide sensitive detection of S-nitrosylated and other modified cysteine residues [110]. Isotope-coded affinity tag (ICAT) approaches allow quantitative assessment of cysteine oxidation states, while novel chemoselective probes enable specific labeling of sulfenic acid modifications [109]. These methodologies require careful control of sample preparation to preserve native redox states and prevent artificial oxidation during extraction.
Quantitative Live-Cell Imaging of Redox Dynamics
Genetically encoded biosensors enable real-time monitoring of redox parameters in living cells. Redox-sensitive GFP (roGFP) coupled to glutaredoxin or ORP1 provides specific measurement of glutathione redox potential, while HyPer probes detect hydrogen peroxide dynamics with high temporal and spatial resolution [70]. Recent advances include semisynthetic biosensors that can be flexibly conjugated to target proteins using Halo- or SNAP-tags, enabling compartment-specific redox monitoring [70].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagents for Redox Biology Studies
| Reagent/Category | Specific Examples | Function/Application | Considerations |
|---|---|---|---|
| Thiol-labeling probes | IAM, NEM, CPM, BIAM | Alkylation of reduced thiols | Must control pH and timing |
| Oxidant-sensitive dyes | Hâ‚‚DCFDA, DHE, MitoSOX | Detection of ROS | Subject to artifacts; requires controls |
| Redox biosensors | roGFP, HyPer, rxYFP | Live-cell imaging of redox state | Requires calibration and proper targeting |
| Modification-specific antibodies | Anti-GSH, Anti-3-NT | Detection of specific modifications | Variable specificity and sensitivity |
| Redox enzyme inhibitors | Auranofin, DPI, ATZ | Target specific redox systems | Often lack complete specificity |
| Thiol reductants | DTT, TCEP, β-ME | Reduction of disulfide bonds | TCEP more stable than DTT |
| Transgenic model systems | sod deletion mutants, Trx overexpression | Genetic manipulation of redox systems | Compensatory mechanisms may develop |
Integrated Cross-Species Analysis: Conserved Principles and Mechanisms
Cysteine Redox Switches as Evolutionarily Conserved Regulatory Nodes
Comparative analysis reveals striking conservation of cysteine redox switches across phylogenetic boundaries. Peroxiredoxins exemplify this conservation, serving as both peroxide scavengers and redox-dependent chaperones while undergoing reversible oxidation cycles regulated by sulfiredoxins [108] [110]. The thioredoxin system similarly demonstrates conserved functionality, with TRX domains maintaining redox regulation of fundamental metabolic enzymes including glyceraldehyde-3-phosphate dehydrogenase and ribonucleotide reductase.
Systems Biology Approaches to Redox Regulation
Constraint-based modeling, including flux balance analysis (FBA), provides powerful computational frameworks for understanding redox network properties. FBA coupled with carbon metabolite tracing revealed unexpected contributions of serine-driven one-carbon metabolism to NADPH production, comparable to the pentose phosphate pathway [70]. These approaches enable prediction of metabolic bottlenecks and identification of critical control points in redox homeostasis.
Figure 3: Experimental Workflow for Redox Proteomics. This diagram outlines integrated approaches for comprehensive analysis of cysteine redox modifications, from sample preparation through systems-level modeling and therapeutic applications.
Therapeutic Implications and Translational Potential
The conserved nature of redox regulatory mechanisms identified in plants and C. elegans provides valuable insights for human disease intervention. Small molecule targeting of specific cysteine residues in redox-sensitive proteins shows promise for selective modulation of pathological processes without disrupting global redox homeostasis [47]. NADPH oxidases (NOX isoforms) represent particularly attractive targets, as their tissue-specific expression and regulated ROS production contribute to signaling in cardiovascular, neurological, and metabolic diseases.
Future Directions and Concluding Perspectives
The integration of plant and C. elegans redox biology continues to reveal fundamental principles of cellular regulation with broad implications for understanding aging and disease. Emerging areas include the role of redox modifications in epigenetic regulation, the coordination of organellar redox communication, and the development of quantitative models predicting redox network behavior under physiological and pathological conditions.
Future research should prioritize spatial mapping of redox modifications within cellular compartments, single-cell redox analysis, and dynamic modeling of redox network flux. The development of more specific redox-based therapeutics will require advanced understanding of context-dependent redox signaling, particularly the distinctions between oxidative eustress and distress. Cross-species comparative approaches will continue to provide unique insights into both conserved and specialized adaptations of redox regulatory systems.
The investigation of cysteine-based redox regulation represents a paradigm for understanding how chemical modifications integrate environmental and metabolic information to control biological fate decisions. The complementary strengths of plant and C. elegans models provide powerful experimental systems for elucidating these mechanisms and translating them into therapeutic strategies for age-related diseases.
The therapeutic targeting of redox-sensitive cysteine residues represents a frontier in precision medicine, bridging the gap between fundamental redox biology and clinical treatment strategies. This in-depth technical guide examines the bench-to-bedside translation of emerging small-molecule inhibitors that selectively target cysteine residues in proteins central to redox signaling pathways. By evaluating mechanistic insights, chemical biology tools, and therapeutic applications across diseases including cancer, metabolic disorders, and cardiovascular conditions, this review provides researchers and drug development professionals with a comprehensive framework for developing redox-based therapeutics. The content emphasizes the sophisticated molecular mechanisms underlying cysteine redox modifications, the current landscape of targeted covalent inhibitors, experimental methodologies for validation, and the challenges in clinical translation. Within the broader context of redox regulation research, this analysis demonstrates how the strategic manipulation of specific cysteine residues offers unprecedented opportunities for therapeutic intervention while highlighting the critical considerations necessary for successful clinical implementation.
Redox regulation of protein cysteine residues has emerged as a fundamental mechanism controlling cellular signaling pathways, with dysregulation implicated in numerous pathological conditions. The term "redox" describes reduction-oxidation chemical reactions involving electron transfer between reactants, processes that are fundamental to biological energy production and signaling networks [47]. Within this framework, cysteine residues serve as critical redox sensors due to the high reactivity of their thiol (-SH) groups, which can undergo reversible oxidative modifications in response to reactive oxygen species (ROS), reactive nitrogen species (RNS), and reactive sulfur species (RSS) [14] [47]. These modifications including sulfenylation (S-OH), S-nitrosylation (S-NO), S-glutathionylation (S-SG), and persulfidation (S-SH) function as molecular switches that dynamically regulate protein function, localization, and interactions [47].
The therapeutic targeting of redox-sensitive cysteines represents a paradigm shift in drug discovery, moving beyond traditional antioxidant approaches toward precise modulation of specific redox nodes in pathological signaling networks. This strategy leverages the unique chemical properties of cysteine residues and their disease-associated modifications to develop targeted covalent inhibitors (TCIs) with enhanced specificity and therapeutic potential [6]. The clinical relevance of this approach is underscored by the approval of covalent kinase inhibitors such as afatinib and ibrutinib, which target specific cysteine residues (Cys797 in EGFR and Cys215 in BTK, respectively), demonstrating the feasibility of cysteine-targeted therapy [6].
This review examines the progression from fundamental research discoveries to clinical application of small-molecule inhibitors targeting redox-sensitive cysteines, addressing the key scientific principles, technological advances, and translational challenges in this emerging field. By integrating recent advances in chemical biology, proteomics, and structural biology, we provide a comprehensive resource for researchers and drug developers working at the intersection of redox biology and therapeutic discovery.
Cysteine Redox Chemistry and Biological Significance
Fundamental Cysteine Redox Modifications
Cysteine residues undergo a spectrum of oxidative modifications that form the molecular basis of redox signaling. The susceptibility of specific cysteine residues to oxidation is determined by multiple factors including local protein microenvironment, pKa of the thiol group, and accessibility to oxidants. The nucleophilic thiolate anion (-S-), which exists at physiological pH when cysteine resides in basic or positively charged environments, is significantly more reactive than the protonated thiol form [47]. This differential reactivity establishes inherent specificity in cysteine oxidation.
The oxidative modification cascade begins with the formation of cysteine sulfenic acid (S-OH), a labile intermediate that serves as a crucial sensor and amplifier of hydrogen peroxide (H2O2) signals [6]. This primary oxidation product can then progress to several reversible modifications:
- Disulfide bonds (S-S): Formed between neighboring cysteine residues or with small thiol compounds like glutathione
- S-glutathionylation (SSG): Mixed disulfides with glutathione that regulate protein function under oxidative stress
- S-nitrosylation (SNO): Addition of nitric oxide derivatives to cysteine thiols
- Persulfidation (SSH): Formation of hydropersulfide groups, part of the reactive sulfur species (RSS) axis [14] [47]
Under severe or persistent oxidative stress, cysteine oxidation can progress to irreversible forms including sulfinic (SO2H) and sulfonic (SO3H) acids, which are typically associated with pathological damage rather than signaling [114]. The identification of serum albumin cysteine trioxidation as a biomarker in type 2 diabetes mellitus exemplifies the clinical relevance of irreversible cysteine oxidation in human disease [114].
Redox-Sensitive Signaling Nodes and Pathways
Specific cysteine residues in regulatory proteins function as critical control points in cellular signaling networks. Key redox-sensitive pathways with therapeutic relevance include:
NRF2-KEAP1 Pathway: The transcription factor NRF2 (nuclear factor erythroid 2-related factor 2) is a master regulator of antioxidant responses, controlling the expression of genes involved in glutathione synthesis, ROS detoxification, and xenobiotic metabolism [115] [47]. Under basal conditions, NRF2 is bound by its negative regulator KEAP1, which contains multiple redox-sensitive cysteine residues (Cys151, Cys273, and Cys288) that function as sensors for electrophiles and oxidants [115]. Modification of these cysteines disrupts the KEAP1-NRF2 interaction, leading to NRF2 stabilization and transcriptional activation of cytoprotective genes.
Ras GTPases: The small GTPases H-Ras, N-Ras, and K-Ras contain a conserved redox-sensitive motif (Asn116-Lys117-Cys118-Asp119) where oxidation of Cys118 affects nucleotide exchange, protein stability, and membrane localization [116]. Multiple oxidative modifications at this site including S-nitrosylation and S-glutathionylation have been documented, with implications for Ras signaling in cancer and cardiovascular disease [116].
Protein Kinase C Isoforms: Multiple PKC isoforms undergo redox-dependent regulation through both allosteric mechanisms and direct cysteine oxidation. PKCδ activation during oxidative stress involves Src-dependent tyrosine phosphorylation, demonstrating integration of redox and phosphorylation signaling networks [117].
Table 1: Clinically Relevant Redox-Sensitive Cysteine Residues
| Protein | Cysteine Residue | Redox Modification | Biological Function | Disease Association |
|---|---|---|---|---|
| KEAP1 | Cys151 | Sulfenylation, Electrophile adduction | NRF2 repression release | Cancer, inflammatory diseases [115] |
| H-Ras | Cys118 | S-nitrosylation, S-glutathionylation | Nucleotide exchange regulation | Cancer, cardiovascular disease [116] |
| EGFR | Cys797 | Irreversible inhibition | Kinase activity | Cancer (afatinib target) [6] |
| BTK | Cys215 | Irreversible inhibition | Kinase activity | B-cell malignancies (ibrutinib target) [6] |
| Human Serum Albumin | Cys34 | Trioxidation (SO3H) | Redox buffer capacity | Type 2 diabetes biomarker [114] |
| PTP1B | Cys215 | Sulfenylation | Phosphatase activity regulation | Diabetes, obesity [47] |
The following diagram illustrates the major cysteine redox modifications and their relationships in signaling and damage contexts:
Diagram 1: Cysteine Redox Modification Pathways. Reversible modifications (green) facilitate signaling, while irreversible oxidation (red) represents oxidative damage.
Emerging Small-Molecule Inhibitors Targeting Redox-Sensitive Cysteines
Chemical Strategies for Targeting Redox-Sensitive Cysteines
The development of small-molecule inhibitors targeting redox-sensitive cysteines employs sophisticated chemical approaches designed to capitalize on the unique reactivity of these residues. Two primary strategies have emerged for therapeutic targeting:
Targeted Covalent Inhibitors (TCIs): These compounds utilize weakly electrophilic warheads that selectively react with nucleophilic cysteine thiols in target proteins. The design follows a two-step mechanism: initial reversible recognition and binding within the target protein's active site, followed by covalent bond formation between the warhead and a proximal cysteine residue [6]. Successful examples include the FDA-approved inhibitors afatinib (EGFR Cys797) and ibrutinib (BTK Cys215), which demonstrate the clinical viability of this approach.
Redox-Based Probe Molecules: Chemical probes designed to mimic or disrupt specific cysteine redox modifications represent an emerging strategy. These include dimedone-based compounds that selectively label sulfenic acid modifications, photocaged cysteine sulfoxide analogs for controlled SOH formation, and persulfide-donating molecules that modulate protein persulfidation [6]. While primarily research tools currently, these probes establish principles for future therapeutic development.
The therapeutic window for redox-targeted inhibitors depends critically on selective targeting of pathological cysteine modifications without disrupting physiological redox signaling. This requires precise tuning of warhead reactivity and optimized binding interactions to achieve target specificity.
Key Inhibitor Classes and Their Targets
Table 2: Emerging Small-Molecule Inhibitors Targeting Redox-Sensitive Cysteines
| Inhibitor Class | Molecular Target | Cysteine Residue | Mechanism of Action | Development Stage |
|---|---|---|---|---|
| Cyanoenone triterpenoids (CDDO derivatives) | KEAP1 | Cys151, Cys273, Cys288 | KEAP1 modification, NRF2 activation | Clinical trials (Bardoxolone methyl) [115] |
| Dimethyl fumarate (DMF) | KEAP1 | Cys151 | Electrophilic modification, NRF2 activation | FDA-approved (multiple sclerosis) [115] |
| Nitroacetamide-based TCIs | Undisclosed sulfenic acid sites | Specific SOH residues | Selective blockade of SOH modifications | Preclinical [6] |
| Ras inhibitors | K-Ras G12C | Mutated Cys12 | Nucleotide pocket trapping, GTPase inhibition | FDA-approved (Sotorasib) [116] |
| AP39 | Mitochondrial proteins | Persulfidation targets | Mitochondria-targeted H2S donation | Preclinical [14] |
| GYY4137 | Multiple protein targets | Persulfidation targets | Slow-release H2S donation | Preclinical [14] |
KEAP1-NRF2 Pathway Activators: Electrophilic compounds that modify KEAP1 cysteine sensors represent a promising class of indirect antioxidants. Bardoxolone methyl, a cyanoenone triterpenoid, modifies specific KEAP1 cysteines (particularly Cys151) to stabilize NRF2 and activate antioxidant gene expression [115]. Similarly, dimethyl fumarate (DMF), approved for multiple sclerosis treatment, activates NRF2 through KEAP1 cysteine modification [115]. Clinical development of these compounds has demonstrated both promise and challenges, with bardoxolone methyl showing efficacy in chronic kidney disease but facing complications in certain patient populations.
Ras Pathway Inhibitors: The discovery of cysteine mutations in Ras GTPases (particularly K-Ras G12C) has enabled development of allele-specific inhibitors. These compounds exploit the nucleophilic thiol of mutated Cys12 to form irreversible covalent bonds that trap Ras in inactive states [116]. Sotorasib received FDA approval for NSCLC with K-Ras G12C mutation, representing a breakthrough in targeting previously "undruggable" oncogenes.
Reactive Sulfur Species Modulators: Donor molecules that release hydrogen sulfide (H2S) or polysulfides represent an emerging approach to modulate protein persulfidation. Compounds including AP39 (mitochondria-targeted) and GYY4137 (slow-release) manipulate the RSS axis to influence metabolic, inflammatory, and epigenetic pathways [14]. Clinical development of H2S donors such as SG1002 has progressed to Phase II trials (NCT02278276), demonstrating translation potential [14].
Experimental Methodologies and Research Tools
Analytical Approaches for Redox Modification Detection
Advancements in analytical technologies have been instrumental in progressing the field of cysteine redox biology toward therapeutic applications. Key methodologies include:
Mass Spectrometry-Based Proteomics: Liquid chromatography-tandem mass spectrometry (LC-MS/MS) platforms enable comprehensive mapping of cysteine modifications across the proteome. Discovery-based data-dependent acquisition (DDA) identifies novel redox-sensitive sites, while targeted approaches like multiple reaction monitoring (MRM) enable precise quantification of specific modifications [114]. The application of these methods identified irreversible cysteine trioxidation on human serum albumin (HSA) at Cys34 as a biomarker of oxidative stress in type 2 diabetes mellitus [114].
Chemoselective Probes and Chemical Proteomics: Bioorthogonal probes that selectively label specific cysteine oxidation states enable enrichment and detection of low-abundance redox modifications. Key probes include:
- Dimedone and derivatives: Selective labeling of sulfenic acids
- Iodoacetyl tandem mass tag (iodoTMT): Quantitative thiol reactivity profiling
- "Tag-switch" reagents: Selective detection of protein persulfidation [14] [6]
These chemoproteomic approaches facilitate system-wide analysis of cysteine redox states under physiological and pathological conditions, identifying novel drug targets and biomarkers.
Functional Validation Techniques
Establishing causal relationships between specific cysteine modifications and functional outcomes requires sophisticated validation approaches:
Genetic Code Expansion for Site-Specific Redox Manipulation: Integration of bioorthogonal cleavage chemistry with genetic code expansion enables precise incorporation of redox-sensitive unnatural amino acids (UAAs) into proteins of interest [6]. For example, photocaged cysteine sulfoxide analogs (e.g., DMNB-caged cysteine sulfoxide) can be site-specifically incorporated, with subsequent UV irradiation generating SOH modifications at defined positions [6]. This approach enables controlled activation of redox events without global oxidative perturbation.
Redox-Targeted Covalent Inhibitor Validation: Comprehensive assessment of TCI specificity and efficacy requires orthogonal approaches:
- Cellular thermal shift assays (CETSA) to confirm target engagement
- Competitive activity-based protein profiling (ABPP) to assess proteome-wide selectivity
- Functional assays measuring pathway modulation and phenotypic outcomes
The following diagram illustrates an integrated experimental workflow for target validation and inhibitor development:
Diagram 2: Experimental Workflow for Redox-Targeted Inhibitor Development
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Redox Cysteine Research
| Reagent Category | Specific Examples | Primary Function | Technical Considerations |
|---|---|---|---|
| Chemoselective Probes | Dimedone, DYn-2, BTD, Cy5-ABP | Sulfenic acid detection and enrichment | Specificity confirmation required; may miss low-abundance sites [6] |
| Unnatural Amino Acids | DMNB-caged cysteine sulfoxide | Site-specific SOH incorporation via genetic code expansion | Requires orthogonal synthetase/tRNA pair; UV decaging may generate ROS [6] |
| RSS Donors | AP39, GYY4137, SG1002 | Modulation of protein persulfidation | Release kinetics and subcellular targeting critical for specificity [14] |
| ROS-Generating Systems | NADPH oxidase constructs, X/XO | Controlled ROS production in cellular models | Physiological vs. pathological levels distinction essential [47] |
| Thiol Blockers | Iodoacetamide, N-ethylmaleimide | Alkylation of reduced thiols prior to redox analysis | Sample preparation standardization required to prevent artifacts [114] |
| Redox Biosensors | roGFP, HyPer | Real-time monitoring of redox dynamics in live cells | Compartment-specific targeting informs microdomain signaling [47] |
Bench-to-Bedside Translation: Challenges and Opportunities
Preclinical Development Considerations
The translation of redox-targeted inhibitors from basic research to clinical application faces several unique challenges:
Therapeutic Window Optimization: Redox signaling operates within narrow physiological ranges, creating a fundamental challenge in achieving therapeutic effects without disrupting essential signaling functions. Compounds must be optimized to target pathological oxidative modifications while preserving physiological redox homeostasis [47]. Dose optimization requires careful consideration of tissue-specific redox states and compensation mechanisms.
Biomarker-Driven Development: The efficacy assessment of redox-targeted therapies necessitates development of specific biomarkers that can:
- Identify patient populations with dysregulated redox pathways
- Monitor target engagement and pathway modulation
- Assess therapeutic efficacy and potential resistance mechanisms
Examples include quantification of irreversible cysteine trioxidation on serum albumin as demonstrated in type 2 diabetes [114], and monitoring of NRF2 activation through target gene expression [115].
Species-Specific Redox Biology: Differences in redox regulation between model organisms and humans present translation challenges. The species-specific expression and regulation of redox sensors, antioxidant enzymes, and metabolic pathways must be considered in preclinical development.
Clinical Translation Landscape
Several redox-targeted approaches have advanced through clinical development, providing valuable insights into translation challenges:
NRF2 Activators in Chronic Disease: Bardoxolone methyl demonstrated significant improvement in kidney function in patients with chronic kidney disease and type 2 diabetes in Phase III trials, but development was complicated by increased cardiovascular adverse events in specific subpopulations [115]. This highlights the critical importance of patient stratification and careful safety monitoring for redox-targeted therapies.
Metabolic and Cardiovascular Applications: Reactive sulfur species modulators have advanced to clinical trials, with SG1002 (a hydrogen sulfide prodrug) demonstrating improved functional capacity and redox status in heart failure patients in Phase II trials [14]. These approaches aim to leverage the vasodilatory, anti-inflammatory, and cytoprotective effects of RSS signaling.
Oncological Applications: The success of K-Ras G12C inhibitors represents a landmark achievement in targeting mutationally activated cysteine residues [116]. Combination approaches with conventional therapies and management of resistance mechanisms represent active areas of investigation.
The targeted manipulation of redox-sensitive cysteine residues represents a promising frontier in precision medicine, offering novel approaches to diseases with significant oxidative stress components. The continued advancement of this field requires interdisciplinary collaboration across chemical biology, proteomics, structural biology, and clinical medicine.
Key future directions include:
- Development of isoform-selective redox modulators to target specific pathway components
- Advanced delivery systems for tissue- and subcellular compartment-specific targeting
- Integration of multi-omics approaches for comprehensive redox network analysis
- Biomarker-driven clinical trial designs for patient stratification and target engagement assessment
As our understanding of the sophisticated language of cysteine redox signaling continues to evolve, so too will our ability to precisely manipulate these pathways for therapeutic benefit. The successful translation of emerging small-molecule inhibitors targeting redox-sensitive cysteines will depend on maintaining a delicate balance between therapeutic efficacy and preservation of physiological redox homeostasis, ultimately fulfilling the promise of redox-based precision medicine.
Conclusion
The redox regulation of protein cysteine residues represents a fundamental language of cellular communication, integral to health and disease. The integration of advanced redox proteomics with AI-driven computational models is rapidly transforming our ability to decode this complex signaling network, moving the field from descriptive biology to predictive science. Key takeaways confirm that specific cysteine modifications act as functional switches controlling critical processes from metabolism to genomic integrity. The future of redox medicine lies not in non-specific antioxidant approaches, but in the precise targeting of key redox-sensitive nodes, such as SRXN1, to re-establish homeostasis. The continued validation of these targets and the development of context-specific therapeutics hold immense promise for mitigating a wide spectrum of human diseases, including metabolic disorders, cancer, and age-related degeneration, heralding a new era of targeted redox-based pharmacotherapy.
References
- 1. The Basics of Thiols and Cysteines in Redox Biology and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4355186/]
- 2. Thiol-Based Redox Switches and Gene Regulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3113447/]
- 3. Thiol-Based Redox Regulation: Key Mechanisms in Plant ... [https://www.frontiersin.org/research-topics/62180/thiol-based-redox-regulation-key-mechanisms-in-plant-growth-and-stress-responseundefined]
- 4. The role of cysteine residues as redox-sensitive regulatory ... [https://pubmed.ncbi.nlm.nih.gov/15582391/]
- 5. The basics of thiols and cysteines in redox biology and ... [https://www.sciencedirect.com/science/article/abs/pii/S0891584914013768]
- 6. Towards site-specific manipulation in cysteine-mediated redox ... [https://pubs.rsc.org/en/content/articlehtml/2025/sc/d5sc02016f]
- 7. Redox States of Protein Cysteines in Pathways of Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7298344/]
- 8. An emerging role for cysteine-mediated redox signaling in ... [https://pubmed.ncbi.nlm.nih.gov/40896924/]
- 9. Cysteine in Longevity-Related Redox Signaling [https://www.fightaging.org/archives/2025/09/cysteine-in-longevity-related-redox-signaling/]
- 10. thiol-based redox regulation drives fungal pathogenesis [https://www.sciencedirect.com/science/article/abs/pii/S0966842X25001799]
- 11. Oxidative stress-mediated protein sulfenylation in human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11402764/]
- 12. A Narrative Review of the Role of S-Glutathionylation in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11944925/]
- 13. S-Persulfidation: Chemistry, Chemical Biology, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7583347/]
- 14. Reactive Sulfur Species and Protein Persulfidation [https://www.mdpi.com/1467-3045/47/9/765]
- 15. S-glutathionylation in cancer: from fundamental ... [https://www.sciencedirect.com/science/article/pii/S1043661825003494]
- 16. Protein S-sulfenylation is a fleeting molecular switch that ... [https://www.nature.com/articles/ncomms12490]
- 17. Nitrosation and nitrosylation [https://en.wikipedia.org/wiki/Nitrosation_and_nitrosylation]
- 18. Nitrosation - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/chemistry/nitrosation]
- 19. Evidence of endogenously produced hydrogen sulfide ... [https://www.nature.com/articles/s41598-022-15360-x]
- 20. BioSemAF-BiLSTM: a protein sequence feature extraction ... [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2025.1616880/full]
- 21. Redox regulation by sulfiredoxin-1: bridging cysteine ... [https://www.nature.com/articles/s12276-025-01563-5]
- 22. Overview of Peroxiredoxins in oxidant defense and redox ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3156475/]
- 23. Overview on Peroxiredoxin - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4749868/]
- 24. Peroxiredoxin [https://en.wikipedia.org/wiki/Peroxiredoxin]
- 25. The thioredoxin system: Balancing redox responses in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10100330/]
- 26. Thioredoxin [https://en.wikipedia.org/wiki/Thioredoxin]
- 27. 140809 - Gene ResultSRXN1 sulfiredoxin 1 [ (human)] [https://www.ncbi.nlm.nih.gov/gene/140809]
- 28. Thioredoxin (Trx): A redox target and modulator of cellular ... [https://www.sciencedirect.com/science/article/pii/S2213231724000089]
- 29. Redox regulation by sulfiredoxin-1: bridging cysteine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12586646/]
- 30. The NRF2-Dependent Transcriptional Regulation of ... [https://www.mdpi.com/2076-3921/11/1/8]
- 31. Transcriptional regulation of the AP-1 and Nrf2 target gene ... [https://pubmed.ncbi.nlm.nih.gov/19326073/]
- 32. Regulation of Nrf2- and AP-1-mediated gene expression ... [https://www.nature.com/articles/aps2010147]
- 33. Transcriptional Regulation by Nrf2 - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC6208165/]
- 34. NRF2 Cysteine Residues Are Critical for Oxidant ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2784728/]
- 35. Distinct Cysteine Residues in Keap1 Are Required for Keap1 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC262403/]
- 36. Transcriptional Regulation of the AP-1 and Nrf2 Target ... [https://www.sciencedirect.com/science/article/pii/S1016847823136430]
- 37. Nrf2 Activation and Antioxidant Properties of Chromone ... [https://www.mdpi.com/1420-3049/30/9/2048]
- 38. Nrf2/Bach1 signaling axis: A promising multifaceted ... [https://www.sciencedirect.com/science/article/pii/S1878747925000649]
- 39. Redox regulation: mechanisms, biology and therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11885647/]
- 40. An emerging role for cysteine-mediated redox signaling in aging [https://pmc.ncbi.nlm.nih.gov/articles/PMC12433492/]
- 41. Dysregulated Redox Signaling and Its Impact on ... [https://www.mdpi.com/2076-3921/14/11/1278]
- 42. Cysteine redoxome landscape in the liver of male mice fed ... [https://www.sciencedirect.com/science/article/pii/S0021925825025827]
- 43. Site-specific mapping and quantification of protein S- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4167403/]
- 44. Redox signaling in innate immunity and inflammation [https://www.sciencedirect.com/science/article/pii/S0891584925007555]
- 45. A redox switch in p21-CDK feedback during G2 phase ... [https://www.sciencedirect.com/science/article/pii/S109727652500646X]
- 46. Integrating Redox Proteomics and Computational Modeling to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12295296/]
- 47. Redox regulation: mechanisms, biology and therapeutic ... [https://www.nature.com/articles/s41392-024-02095-6]
- 48. Contemporary proteomic strategies for cysteine redoxome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8154054/]
- 49. Redox Proteomics: A Key Tool for New Insights into Protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5327050/]
- 50. Proteome-wide analysis of cysteine oxidation reveals ... [https://www.nature.com/articles/s41467-018-04003-3]
- 51. Deciphering protein long-chain S -acylation using mass ... [https://pubs.rsc.org/en/content/articlehtml/2025/cb/d5cb00146c]
- 52. Proteomic Approaches to Study Cysteine Oxidation [https://www.frontiersin.org/journals/molecular-neuroscience/articles/10.3389/fnmol.2021.678837/full]
- 53. Integrating Redox Proteomics and Computational ... [https://www.mdpi.com/1422-0067/26/14/6925]
- 54. Integrating Redox Proteomics and Computational ... [https://pubmed.ncbi.nlm.nih.gov/40725172/]
- 55. IodoTMT-labeled redox proteomics reveals the ... [https://pubmed.ncbi.nlm.nih.gov/37224778/]
- 56. Quantitative proteomics identifies redox switches for global ... [https://www.nature.com/articles/s41467-017-02694-8]
- 57. Targeted Quantitation of Site-Specific Cysteine Oxidation in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2938085/]
- 58. Proteome-wide analysis of cysteine oxidation reveals ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5910380/]
- 59. oxSWATH: An integrative method for a comprehensive redox ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6435957/]
- 60. CysQuant: Simultaneous quantification of cysteine ... [https://pubmed.ncbi.nlm.nih.gov/37793239/]
- 61. simultaneous quantification of cysteine oxidation and ... [https://www.omicsdi.org/dataset/pride/PXD043895]
- 62. pCysMod: Prediction of Multiple Cysteine Modifications ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7959776/]
- 63. Redox proteomics combined with proximity labeling ... [https://www.sciencedirect.com/science/article/pii/S2451945623000557]
- 64. Metabolomics and transcriptomics based multi-omics ... [https://www.nature.com/articles/s41540-023-00305-5]
- 65. Strategies for Comprehensive Multi-Omics Integrative Data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11592251/]
- 66. Convergence: Multi-omics and AI are reshaping the ... [https://www.sciencedirect.com/science/article/abs/pii/S0925443925003758]
- 67. Cysteine redoxome landscape in the liver of male mice fed ... [https://pubmed.ncbi.nlm.nih.gov/40967437/]
- 68. Cysteine redoxome landscape in the liver of male mice fed a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12547247/]
- 69. Cysteine redoxome landscape in mouse brown adipose ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11915156/]
- 70. Redox Systems Biology: Harnessing the Sentinels of the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7047077/]
- 71. Quantitative analysis of redox proteome reveals oxidation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9157258/]
- 72. Thiol-based Oxidative Posttranslational Modifications ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9282725/]
- 73. Global profiling of distinct cysteine redox forms reveals ... [https://www.nature.com/articles/s41467-021-21686-3]
- 74. Auxin-Mediated Redox Control of the Ubiquitin ... [https://www.techscience.com/biocell/v49n10/64056/html]
- 75. Cysteine Reactivity Across the Sub-Cellular Universe - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6382561/]
- 76. Quantitative reactivity profiling predicts functional cysteines ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3058684/]
- 77. The Local Electrostatic Environment Determines Cysteine ... [https://www.sciencedirect.com/science/article/pii/S0021925820703108]
- 78. CysDB: a human cysteine database based on ... [https://www.sciencedirect.com/science/article/pii/S2451945623000909]
- 79. muscle Compartmentalized controlling exercise... redox signals [https://pmc.ncbi.nlm.nih.gov/articles/PMC7284909/]
- 80. The Power of Compartmentalization : A Biochemical Perspective [https://www.numberanalytics.com/blog/power-of-compartmentalization-biochemical-perspective]
- 81. Protein redox chemistry: post-translational cysteine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4186267/]
- 82. Correlation of organelle interactions in the development ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1567743/full]
- 83. Structural insights into redox-active cysteine residues of the ... [https://www.sciencedirect.com/science/article/pii/S2213231721000823]
- 84. Protein Kinases: Redox takes control [https://elifesciences.org/articles/99765]
- 85. Free radicals and their impact on health and antioxidant ... [https://www.nature.com/articles/s41420-024-02278-8]
- 86. Chemical and molecular mechanisms of antioxidants [https://pmc.ncbi.nlm.nih.gov/articles/PMC2927345/]
- 87. Antioxidants: a comprehensive review [https://link.springer.com/article/10.1007/s00204-025-03997-2]
- 88. Antioxidants and Reactive Oxygen Species [https://www.mdpi.com/1422-0067/26/15/7520]
- 89. 线粒体功能障ç¢ä¸Žå¸•é‡‘森病 [https://biospective.com/zh/resources/mitochondrial-dysfunction-parkinsons-disease]
- 90. Protein oxidation - Formation mechanisms, detection and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8113053/]
- 91. CysQuant: Simultaneous quantification of cysteine oxidation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10562924/]
- 92. Model organisms for investigating the functional ... [https://www.sciencedirect.com/science/article/pii/S2213231724004427]
- 93. H2 as a potential redox homeostasis modulator: a review of ... [https://pubmed.ncbi.nlm.nih.gov/41254431/]
- 94. Kynurenine-mediated redox regulation provides ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-07214-7]
- 95. bridging cysteine oxidation and liver disease therapeutics [https://pubmed.ncbi.nlm.nih.gov/41131335/?utm_source=FeedFetcher&utm_medium=rss&utm_campaign=None&utm_content=0lAgosEFCISvVtB9Gz6iR5ulz5j0-ZrCLAmXoXIH_-e&fc=None&ff=20251028074448&v=2.18.0.post22+67771e2]
- 96. SRXN1 stimulates hepatocellular carcinoma tumorigenesis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7521256/]
- 97. SRXN1 Gene [https://maayanlab.cloud/Harmonizome/gene/SRXN1]
- 98. The desulfinylation enzyme sulfiredoxin-1 attenuates HSC ... [https://pubmed.ncbi.nlm.nih.gov/39446334/]
- 99. Sulfiredoxin as a Potential Therapeutic Target for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7201699/]
- 100. Oxidative Cysteine Post Translational Modifications Drive ... [https://www.mdpi.com/2076-3921/13/8/883]
- 101. Redox Regulation of Protein Function via Cysteine S ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3431077/]
- 102. Redox Post-translational Modifications of Protein Thiols in ... [https://www.frontiersin.org/journals/aging-neuroscience/articles/10.3389/fnagi.2020.00254/full]
- 103. Hydrogen sulfide regulation in redox homeostasis and ... [https://www.sciencedirect.com/science/article/pii/S2090123225007659]
- 104. The Redox Paradox: Cancer's Double-Edged Sword for ... [https://www.mdpi.com/2076-3921/14/10/1187]
- 105. Targeting endoplasmic reticulum stress and nitroso-redox ... [https://www.nature.com/articles/s41420-025-02774-5]
- 106. Exploring the Antioxidant Roles of Cysteine and ... [https://www.mdpi.com/2218-273X/15/8/1115]
- 107. Selenium Speeds Reactions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1278939/]
- 108. Redox regulation in lifespan determination - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10965828/]
- 109. Post-Translational Modifications to Cysteine Residues in ... [https://www.mdpi.com/1422-0067/25/18/9845]
- 110. Cysteine–based redox regulation and signaling in plants [https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2013.00105/full]
- 111. Quantitative Redox Biology: An approach to understanding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3661692/]
- 112. Redox-based protein modifications: the missing link in ... [https://www.sciencedirect.com/science/article/abs/pii/S1369526611000197]
- 113. Modulation of longevity and diapause by redox regulation ... [https://www.sciencedirect.com/science/article/abs/pii/S0531556508000685]
- 114. Serum albumin cysteine trioxidation is a potential oxidative ... [https://www.nature.com/articles/s41598-020-62341-z]
- 115. Health position paper and redox perspectives - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC11970334/]
- 116. Cysteine-based regulation of redox-sensitive Ras small ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6695279/]
- 117. Mechanisms for redox-regulation of protein kinase C - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4477140/]